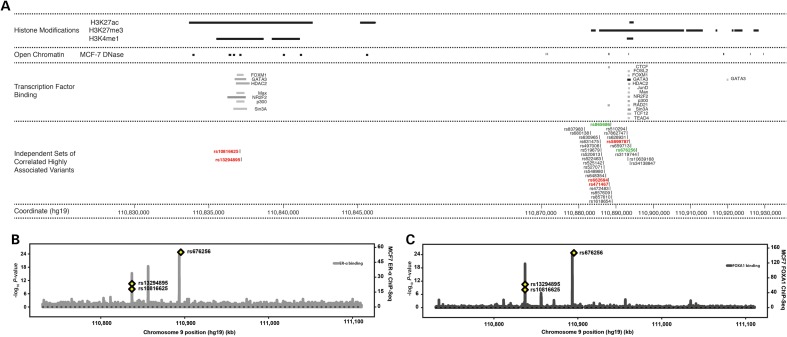
Figure 2.
Plots of genomic annotations with putative functional significance at the 9q31.2 fine-mapping region. (A) Publically available histone modification, DNase hypersensitivity and transcription factor binding data from MCF7 cells were mapped on to the breast cancer associated regions identified by fine-mapping. For SNPs rs10826625 and rs13294895, the iCHAVs were defined as SNPs having r2 ≥ 0.8 with either SNP; for rs676256 it was defined as all SNPs with r2 ≥ 0.8 and likelihood ratios >1:100 relative to rs676256. There were no other SNPs in the iCHAVs for rs10816625 and rs13294895. The rs676256 iCHAV comprised 28 SNPs. SNPs whose identifiers are shown in red type were of putative functional significance (see Materials and Methods). Where the lead SNP was not deemed to be of putative functional significance, it is indicated in green, as is the index 9q31.2 SNP, rs865686. (B) Regional binding profiles for ER-α in MCF7 cells shown plotted across the fine-mapping region using data from (31). The locations of the lead SNPs are indicated with yellow diamonds. (C) Regional binding profiles for FOXA1 in MCF7 cells shown plotted across the fine-mapping region using data from (31). The locations of the lead SNPs are indicated with yellow diamonds.