Figure 3.
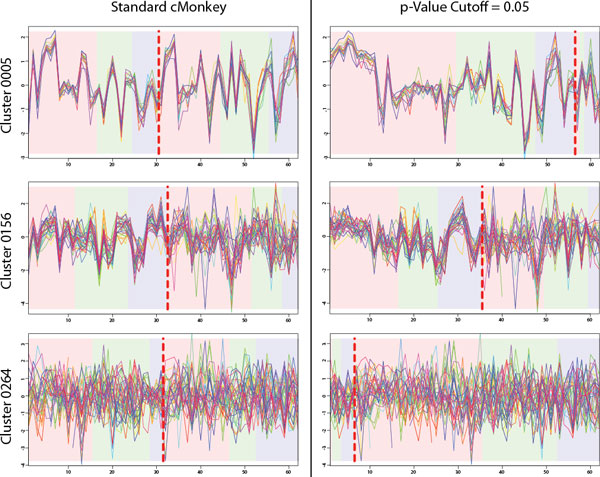
Re-splitting Biclusters Based on BSCM. These biclusters were built on S. cerevisiae grown in batch culture and chemo stat and run through a microarray at different time points. Those conditions to the left of the dotted red line are included in the cluster and those to the right are excluded. The variance based p-value is applied to re-split the biclusters and change which conditions are included. Cluster 0005 contains 15 ribosomal genes that are expected to be co-regulated. Re-splitting increases the number of included conditions from 30/62 to 56/62. Cluster 0156 contains 31 genes related to mitosis, DNA damage, and metabolism. Re-splitting increases the number of included conditions from 32/62 to 35/62. Cluster 0229 contains 34 genes of mostly unknown function although some were related to carbohydrate metabolism. Re-splitting reduces the number of included conditions from 34/62 to 18/62.
