Figure 3.
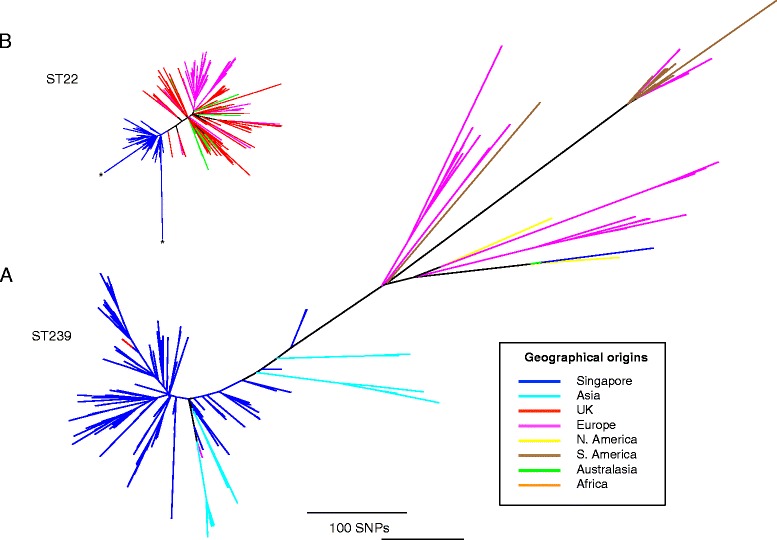
Global context of Singaporean ST22 and ST239 MRSA populations. (A, B) Unrooted maximum likelihood trees of Singaporean ST239 (A) and ST22 (B) isolates including additional isolates representative of the global population of these two lineages. The ST239 phylogeny includes the isolates described by Harris et al. [14] and Castillo-Ramirez et al. [16], and the ST22 phylogeny contains the isolates described by Holden et al. [10]. In the case of ST22, only ST22-A isolates were included as these are representative of EMRSA-15, the clone from which the Singaporean ST22 isolates emerged. Trees were built using a maximum likelihood method using SNPs from the core genome, where the ST22 isolate reads were mapped to the reference chromosome of HO 5096 0412 [20], and the ST239 isolate reads mapped to the TW20 [14] reference chromosome. Branches are colored according to their geographical origins (see legend). The scale bar represents substitutions per SNP site for both trees. The two ST22 isolates excluded from the Bayesian analysis as potential hyper-mutators (B11318/05 and DB055850/09) are marked on the figure by asterisks.
