Figure 5.
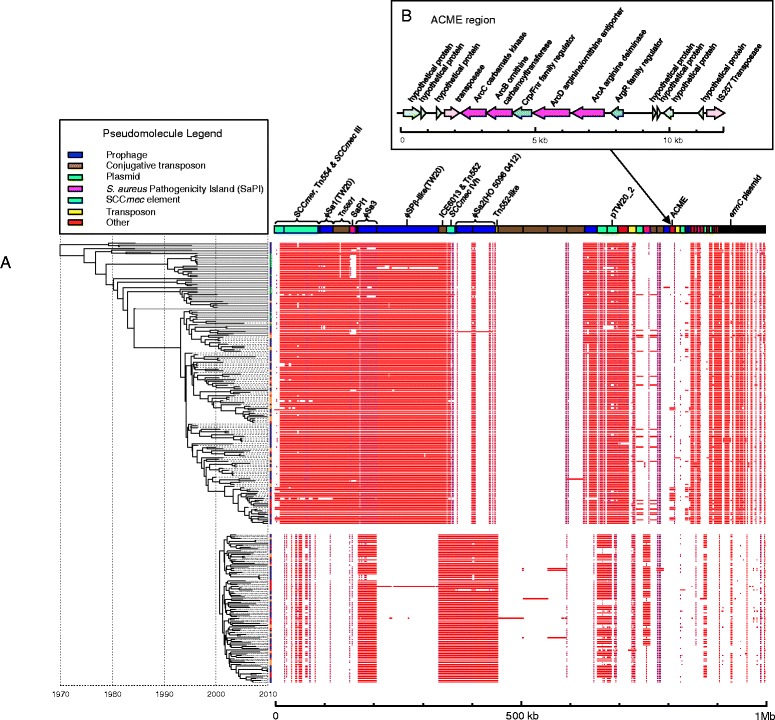
Comparison of the accessory genomes of ST239 and ST22. (A) The presence and absence, by mapping, of components of the accessory genomes of the Singaporean ST239 and ST22 isolates is shown in the panel on the right. For the figure, the colors of the blocks represent percentage identity, going from white, indicating absence or less than 90% identity, through to red (via purple), indicating 100% identity. Contigs of the combined accessory genomes are displayed as a pseudomolecule at the top of the panel, and are colored according to the type of MGE (see legend to figure) they belong to, based on contig annotation and also comparison with an MGE reference set. The contigs are ordered from left to right to compile MGEs from the ST239 reference (TW20), and then the ST22 reference (HO 5096 0412) chromosomes (in the order they appear in the chromosome), then additional accessory genome components (in the order of large to small size). Text above the contigs indicates the reference MGE name, or the name of the accessory elements where found. Isolates are ordered according to the maximum likelihood trees displayed on the left; ST239 isolates are displayed at the top, and ST22 isolates at the bottom. Colored balls on the tips of the tree indicate the origins of the isolate (see Figure 2 for legend). (B) Gene organization of the type II arginine catabolic mobile element identified in the ST239 pan-genome. Coding sequences, indicated by arrows, are colored according to functional groups: pink, transposition; blue, regulation; magenta, catabolism; green, hypothetical protein.
