Figure 2.
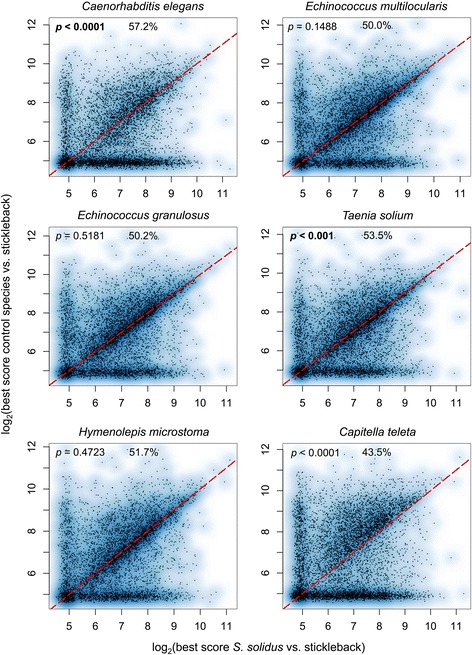
Protein similarity comparisons using full-length blastp searches against various parasitic and non-parasitic control worm species. Scatter plots of the best blastp scores of S. solidus proteome (x axis) and six control worm species proteomes (y axis, name of the species specified on top of each graph) against the proteome of the threespine stickleback. Data below the red dotted line correspond to stickleback proteins showing a higher sequence similarity for S. solidus as compared to the corresponding control worm species. For each scatter plot, a Wilcoxon signed-rank test was performed on the distributions of blastp scores (p-value in the upper left corner of each graph). Significant p-values were highlighted in bold for higher blastp scores between S. solidus and its host than between the control species and the host. The percentage of the stickleback proteome showing higher scores with S. solidus than with the control species appears on top of each scatterplot.
