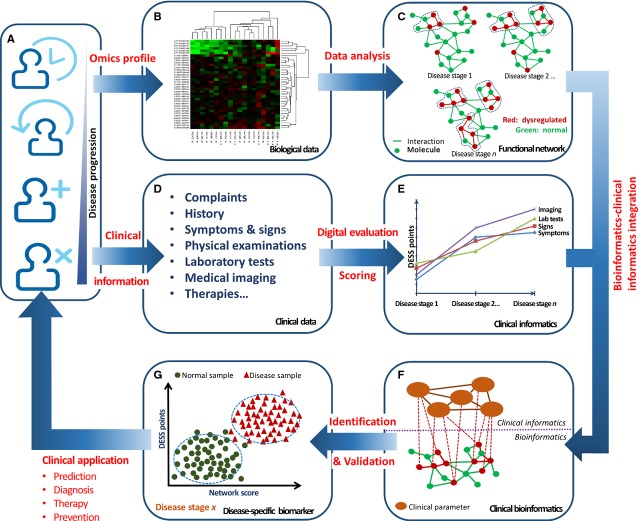
Fig 4.
A proposed workflow of disease-specific biomarker identification by integration of bioinformatics and clinical informatics. Both the global expression profiles (B; genomics, proteomics, etc.) and the clinical data (D) of a disease phenotype at different stages (A) are obtained. Disease-associated functional networks (C) are measured by bioinformatics, while clinical informatics (E) is generated through a digital evaluation score system (DESS). By integrating bioinformatics and clinical informatics, the molecular-phenotype networks are measured using different methods to score, rank and identify candidate biomarkers (F). The identified disease-specific biomarkers are then validated to differentiate a disease phenotype from a normal phenotype (G) for clinical application to develop predictive, diagnostics and preventive methods for personalized medicine.