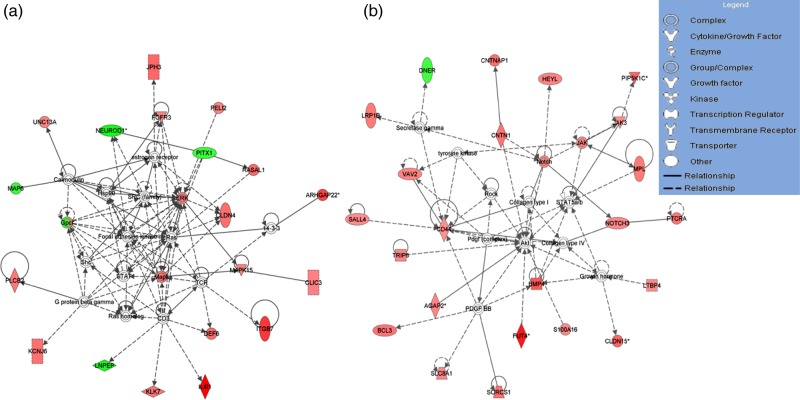
Figure 2.
The two representative networks by ingenuity pathway analysis (IPA) for differentially methylated whole gene set derived from HF-dNK-treated EVT. (a) Extracellular signal-regulated protein kinase 1 and 2 (Erk1/2) was centered in one of the top network pathways, and Claudin-4 (CLDN4) was revolved this pathway. (b) Protein kinase B (AKT) was centered in another top network pathway, and fucosyltransferase IV (FUT4) was revolved this pathway. The analyses indicate that the differentially methylated genes from HF-dNK-treated EVT are located in pathways related to cellular development, cellular function and maintenance, and cell-to-cell signaling and interaction, revolving around key cellular proteins AKT and Erk1/2. The color intensity of node indicates the degree of hyper- or hypo-methylation. Genes encoding for molecules in red are hypermethylated while those in green are hypomethylated relative to control EVT. The shape of each node denotes the molecule class displayed beside each figure. The nodal relationships indicated in solid lines denote direct while those in dashed lines denote indirect interactions. AGAP2: ArfGAP with GTPase domain, ankyrin repeat and PH domain 2; ARC: Activity-regulated cytoskeleton-associated protein; BCL3: B cell lymphoma 3 protein; BMP4: bone morphogenetic protein 4; CD3: cluster of differentiation 3; CD44: cluster of differentiation 44; CLIC3: chloride intracellular channel 3; CNTN1: contactin 1; CNTNAP1: contactin associated protein 1; DEF6: differentially expressed in FDCP 6 homolog; DNER: delta and notch-like epidermal growth factor-related receptor; FGFR3: fibroblast growth factor receptor 3; GPC1: glypican 1; HEYL: hes-related family bHLH transcription factor with YRPW motif-like; Hsp90: 90 kDa heat shock protein; IL4I1: interleukin 4 induced 1; ITGB7: intergrin, beta 7; JAK: janus kinase; JAK3: janus kinase 3; KCNJ6: potassium inwardly-rectifying channel, subfamily J, member 6; KLK7: kallikrein-related peptidase 7; LNPEP: leucyl/cystinyl aminopeptidase; LRP1B: low density lipoprotein receptor-related protein 1B; MAP6: microtubule-associated protein 6; MAPK: mitogen-activated protein kinase; MPL: myeloproliferative leukemia virus oncogene; NEUROD1: neuronal differentiation 1; NOTCH3: Notch homolog 3; PDGF BB: platelet-derived growth factor beta polypeptide homodimer; PELI2: pellino E3 ubiquitin protein ligase family member 2; PIP5K1C: phosphatidylinositol-4-phosphate 5-kinase, type I, gamma; PITX1: paired-like homeodomain 1; PLCB2: phospholipase C, beta 2; PTCRA: pre T-cell antigen receptor alpha; RASAL1: RAS protein activator like 1; ROCK: rho-associated, coiled-coil-containing protein kinase; S100A16: S100 calcium binding protein A16; SALL4: spalt-like transcription factor 4; Shc: SHC (Src homology 2 domain containing) transforming protein; SLC8A1: solute carrier family 8 (sodium/calcium exchanger), member 1; SORCS1: sortilin-related VPS10 domain containing receptor 1; STAT: signal transducer and activator of transcription; STAT5a/b: signal transducer and activator of transcription 5a/b; TCR: T cell receptor; TRIP6: thyroid hormone receptor interactor 6; UNC13A: protein unc-13 homolog A; VAV2: vav 2 guanine nucleotide exchange factor.