Abstract
The α9α10 nicotinic acetylcholine receptor (nAChR) was first identified in the auditory system, where it mediates synaptic transmission between efferent olivocochlear cholinergic fibers and cochlea hair cells. This receptor gained further attention due to its potential role in chronic pain and breast and lung cancers. We previously showed that α-conotoxin (α-CTx) RgIA, one of the few α9α10 selective ligands identified to date, is 300-fold less potent on human versus rat α9α10 nAChR. This species difference was conferred by only one residue in the (−), rather than (+), binding region of the α9 subunit. In light of this unexpected discovery, we sought to determine other interacting residues with α-CTx RgIA. A previous molecular modeling study, based on the structure of the homologous molluscan acetylcholine-binding protein, predicted that RgIA interacts with three residues on the α9(+) face and two residues on the α10(−) face of the α9α10 nAChR. However, mutations of these residues had little or no effect on toxin block of the α9α10 nAChR. In contrast, mutations of homologous residues in the opposing nAChR subunits (α10 Ε197, P200 and α9 T61, D121) resulted in 19- to 1700-fold loss of toxin activity. Based on the crystal structure of the extracellular domain (ECD) of human α9 nAChR, we modeled the rat α9α10 ECD and its complexes with α-CTx RgIA and acetylcholine. Our data support the interaction of α-CTx RgIA at the α10/α9 rather than the α9/α10 nAChR subunit interface, and may facilitate the development of selective ligands with therapeutic potential.
Introduction
Nicotinic acetylcholine (ACh) receptors (nAChRs) are members of the Cys-loop family of ligand-gated ion channels, whose other members include 5HT3, the GABAA, and glycine receptors (Lester et al., 2004; Sine and Engel, 2006; Bouzat, 2012). nAChRs are pentameric receptors found both at the neuromuscular junction as well as in central and peripheral neurons. The neuronal nAChRs are composed of a combination of α and β subunits. To date, nine nonmuscle α (α2–10) and three β (β2–4) subunits have been identified (Albuquerque et al., 2009). Different combinations of α/β (heteromeric) or α/α subunits (homomeric) result in formation of nAChRs with distinct pharmacology and localization within both the central and peripheral nervous system (Gotti et al., 2006, 2007; Millar and Gotti, 2009). One of these subunits, α9, was initially localized in cochlear inner and outer hair cells (Elgoyhen et al., 1994). Although originally thought to form a homomeric α9 nAChR, subsequent studies identified the α10 subunit as a partner (Elgoyhen et al., 2001; Lustig et al., 2001; Sgard et al., 2002). Although similar to homomeric α9 nAChRs, heteromeric α9α10 nAChRs more closely resemble the pharmacological and electrophysiological properties of native nAChRs found in cochlear hair cells (Elgoyhen et al., 2001; Sgard et al., 2002; Lustig, 2006). In addition, both knockout and developmental studies indicate that responses of cochlear hair cells to ACh are mediated through α9α10 nAChRs (Vetter et al., 1999, 2007; Katz et al., 2004; Roux et al., 2011).
In addition to the auditory system, transcripts and/or protein for α9 and α10 subunits have been found in lymphocytes, skin keratinocytes, sperm, dorsal root ganglion, sympathetic neurons, macrophages, and adrenal chromaffin cells (Nguyen et al., 2000; Lustig et al., 2001; Lips et al., 2002; Haberberger et al., 2004; Kurzen et al., 2004; Peng et al., 2004; Kumar and Meizel, 2005; Colomer et al., 2010; Simard et al., 2013). The physiologic role of α9α10 nAChRs in the auditory system has been determined through studies done with α9 and α10 knockout mice (Vetter et al., 1999, 2007). These studies suggest an important role for both subunits in the development of cochlear morphology and innervation, as well as in the regulation of normal suppression of cochlear responses after olivocochlear fiber stimulation (Vetter et al., 1999, 2007).
In contrast to the auditory system, the physiologic role of α9α10 nAChRs in other areas has been more difficult to assess. For example, in human lymphocytes that coexpress mRNA for both α9 and α10 subunits, ACh failed to generate an ionic current, suggesting a physiologic role of α9α10 nAChRs different from that observed in cochlear hair cells (Peng et al., 2004). Recently, it was shown that α-conotoxin (α-CTx) RgIA, a specific blocker of α9α10 nAChRs (Ellison et al., 2006), is analgesic in a rat model of nerve injury (Vincler et al., 2006). This analgesic effect was attributed to a decrease in the number of immune cells recruited to the site of injury (Vincler et al., 2006).
The potency of α-CTx RgIA for heterologously expressed rat versus native α9α10 nAChRs expressed in hair cells matches well (Ellison et al., 2006). However, upon comparing the potency of α-CTx RgIA on heterologously expressed rat versus human α9α10 nAChRs, we observed a >300-fold difference in potency between the two species, with the toxin being less potent on the human subtype. Subsequent mutation studies indicated that this difference is conferred by a single amino acid in the (−) binding region of the α9 subunit (Azam and McIntosh, 2012). In the current study, we have performed an in-depth structure/function analysis of the interaction of α-CTx RgIA with the α9α10 nAChR.
Materials and Methods
ACh chloride, atropine, and bovine serum albumin were obtained from Sigma-Aldrich (St. Louis, MO). α-CTxs were synthesized, as described previously (McIntosh et al., 2005; Ellison et al., 2006). Clones of rat α9 and α10 cDNAs in pGEMHe and pSGEM vectors, respectively, were provided by A. Belen Elgoyhen (Universidad de Buenos Aires, Buenos Aires, Argentina).
Construction of Point Mutations.
Point mutants were made by polymerase chain reaction. Primers containing the desired point mutation flanked by at least 15 bases on either side were synthesized. Using the nonstrand-displacing action of Pfu Turbo DNA polymerase, mutagenic primers were extended and incorporated by polymerase chain reaction. The methylated, nonmutated parental cDNA was digested with DpnI. Mutated DNA was transformed into DH10B or DH5α competent cells, isolated using the Qiagen miniprep kit, and sequenced to ascertain the incorporation of the desired mutation.
cRNA Preparation and Injection.
Capped cRNA for the various subunits were made using the mMessage mMachine in vitro transcription kit (Ambion, Austin, TX) following linearization of the plasmid. cRNA was purified using a Qiagen RNeasy kit (Qiagen, Valencia, CA). The concentration of cRNA was determined by absorbance at 260 nm. cRNA of either wild-type or mutant subunits was mixed at a 1:1 ratio with wild-type subunits for a final concentration of at least 500 ng/μl for each subunit cRNA. One hundred to 150 nl of this mixture was injected into each Xenopus oocyte with a Drummond microdispenser (Drummond Scientific, Broomall, PA), as described previously (Cartier et al., 1996), and incubated at 17°C. Oocytes were injected within 1 day of harvesting, and recordings were made 2–4 days postinjection.
Voltage-Clamp Recording.
For the antagonist dose-response experiments, oocytes were voltage-clamped and exposed to ACh and peptide, as described previously (Cartier et al., 1996). Briefly, the oocyte chamber consisting of a cylindrical well (∼30 μl in volume) was gravity perfused at a rate of ∼2 ml/min with ND-96 buffer (96.0 mM NaCl, 2.0 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 5 mM HEPES, pH 7.1–7.5, supplemented with 0.1 mg/ml bovine serum albumin). In the experiments with Ca2+-free ND96, 1.8 mM BaCl was substituted. Oocytes were subjected once per minute to a 1-second pulse of 100 μM ACh. For toxin concentrations of 1 μM and lower, once a stable baseline was achieved, either ND-96 alone or ND-96 containing varying concentrations of the α-CTxs were perfusion-applied, during which 1-second pulses of 100 μM ACh (200 μM for α7 nAChR) were applied every 90 seconds until a constant level of block was achieved. For toxin concentrations of 10 μM and higher, the buffer flow was stopped and the toxin was bath-applied and allowed to incubate with the oocyte for 5 minutes, after which the ACh pulse was resumed.
To acquire ACh dose-response data, the conventional oocyte chamber was replaced by a chamber constructed from a disposable 200-µl polypropylene pipette tip with a length of 50 mm and an internal diameter of 0.5 mm at the upstream or intake end and 5 mm at the downstream or exhaust end. The chamber was mounted horizontally with its intake end connected to the perfusion supply, whereas its exhaust end had a vertical meniscus whose location was dictated by the tip of a sipper made from a 27-gauge hypodermic needle connected to a vacuum line. The chamber had two apertures in its dorsal wall, as follows: 1) a 1.5-mm circular hole centered 13 mm downstream from the intake, and 2) a 2.5 × 5-mm (longitudinal) oval centered 14 mm downstream from the hole (i.e., a total of 27 mm from the intake end). Oocytes were introduced into the chamber through the oval aperture and secured against the chamber floor by two voltage-clamp glass microelectrodes that impaled the oocyte. The chamber was perfused at a rate of ∼1 ml/min. To introduce ACh into the chamber, the perfusion was halted and 20 µl ACh was manually applied to the chamber via the small circular hole upstream from the oocyte. This volume was too small for ACh to reach the oocyte unless the perfusion was resumed. Upon resumption of perfusion (which was started immediately following the introduction of ACh into the chamber), the bolus of ACh rapidly engulfed the oocyte and washed past it in a matter of seconds, as judged by the time course of ACh response. This process was repeated with different concentrations of ACh with a time interval between applications long enough to avoid desensitization.
Data Analysis.
For the baseline measurement, at least three ACh responses were averaged. To determine the percent block induced by toxin, two to three ACh responses, obtained after a steady state block had been achieved, were averaged and the value divided by the pretoxin baseline value to yield a percentage response. Dose-response data were fit to the equation, Y = 100/(1 (+) 10^((LogEC50 − Log[Toxin]) × nH)), where nH is the Hill coefficient, by nonlinear regression analysis using GraphPad Prism (GraphPad Software, San Diego, CA). Each data point is the mean ± S.E.M. from at least three oocytes. For ACh dose-response curves, the response to a given ACh concentration was normalized to the response to 100 μM ACh, which served as an internal control.
Molecular Modeling Methods.
The molecular model of the extracellular domain (ECD) of the rat (α9)2(α10)3 AChR was based on the high-resolution (1.7 Å) X-ray crystal structure of the monomeric state of the ECD of the human α9 nAChR in its complex with the antagonist methyllycaconitine (PDB ID 4UXU) (Zouridakis et al., 2014). All nonprotein atoms and the alternative location B of residues H63 and N109 with the lowest occupancy were removed from the template structure. The sequence alignment between the human and rat ECDs (Supplemental Fig. 1) was performed with ClustalW2 using the UNIPROT accession codes P43144 for rat α9 (96.2% sequence identity for 212 residues) and Q9JLB5 for rat α10 (66.7% sequence identity). The homology models of the rat α9 and α10 monomers were prepared using Modeler v9.10 (Fiser and Sali, 2003), and the best models were selected on the basis of the lowest discrete optimized protein energy score among 30 models generated. The initial model of the α-CTx RgIA was taken from the representative conformation (model 1) of its NMR structure (PDB ID 2JUQ) (Ellison et al., 2008). The (α9)2(α10)3 ECD was prepared by superimposing each of the two rat monomeric models on the crystallographic structure of the Aplysia californica ACh-binding protein (AChBP) complex with α-CTx ImI (PDB ID 2BYP) (Hansen et al., 2005) using the MULTISEQ plugin of VMD v1.9 (Humphrey et al., 1996). Specifically, α10 ECD was superimposed with chains A, B, and D, whereas α9 ECD was superimposed with chains C and E, so that two α10(+)α9(−) binding sites between chains B, C and D, E were generated. The final model of the rat (α9)2(α10)3 complex with α-CTx RgIA was prepared by superimposing the NMR structure of RgIA with chains F and I of ImI in the AChBP complex. The α9α10P200Q mutant was prepared by changing P200 of a single α10 subunit to glutamine in the final model. The model of the rat (α9)2(α10)3 complex with ACh was prepared by a similar procedure based on the X-ray crystal structure of the Lymnaea stagnalis AChBP complex with carbamylcholine (PDB ID 1UV6) (Celie et al., 2004). Ligand molecules were placed at the five ligand binding sites by changing only the amide nitrogen atom of carbamylcholine to carbon and then adding hydrogen atoms.
Molecular Dynamics Simulations.
Molecular dynamics (MD) simulations were performed using the GPU-PMEMD program of AMBER v12 software (Case et al., 2005; Salomon-Ferrer et al., 2013). The ff12SB force field parameters (Hornak et al., 2006) were employed for the receptor and the α-CTx RgIA, whereas parameters from the General Amber Force Field (Wang et al., 2004) with AM1-BCC charges were applied to the ligands using the LEaP module. The systems were solvated in truncated octahedron boxes of TIP3P waters with a minimum extension of 12 Å from the solute, and the total charge was neutralized with the addition of sodium ions. Simulations were performed with a 2-femtosecond time step and the SHAKE algorithm to constrain all bonds involving hydrogen atoms. The Particle Mesh Ewald method was used for long-range electrostatic interactions, and an 8-Å cutoff radius was used for range-limited interactions. Initially, the system was minimized and then the temperature increased gradually from 10 K to 300 K within 100 picoseconds using harmonic positional restraints of 10 kcal/mol per Å2 on the protein and peptide backbone atoms. The restraints were then gradually removed within 200 picoseconds, and the system was equilibrated for 10 nanoseconds at constant isotropic pressure of 1 atm and temperature of 300 K, using the Berendsen control algorithms. The production simulation was carried out for 100 nanoseconds in the isothermal-isobaric (NPT) ensemble with a pressure relaxation time of 2 picoseconds, using a Langevin thermostat at 300 K with a collision frequency of 2 picoseconds. Snapshots were collected every 2 picoseconds and were processed using the PTRAJ module of AMBER. The structures were clustered using a hierarchical agglomerative approach with a minimum distance between clusters of 1.5 Å, after mass-weighted, root-mean-square deviation fitting of all Cα atoms (Supplemental Material 1 and 2). Calculations were performed on an Intel workstation equipped with NVIDIA GTX 780 GPUs running a Linux 2.6.32 86_x64 kernel.
Results
Identification of Determinants of α-CTx RgIA Interaction with α9 and α10 Subunits
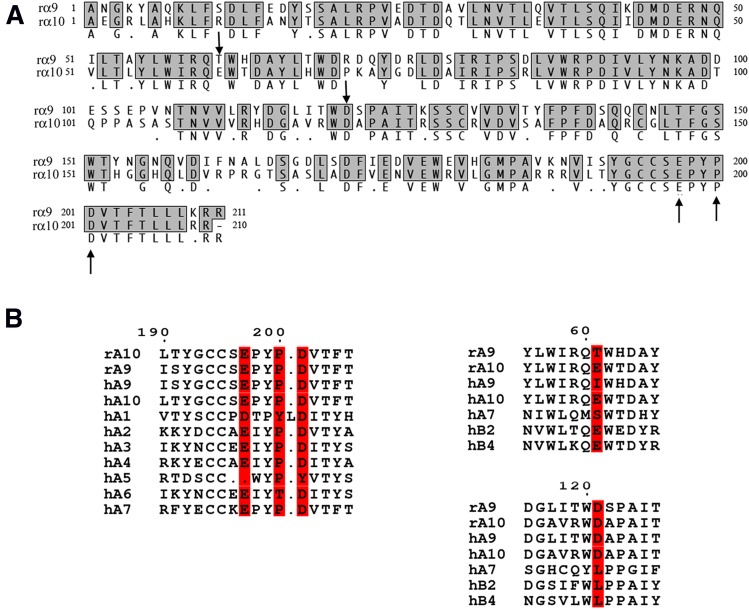
Competitive nicotinic ligands bind to the interface of adjacent nAChR subunits. The α9α10 nAChR may have two or more ligand-binding interfaces that lie at the junction between α9 and α10 subunits. These binding interfaces have historically been believed to occur between the (+) side of α9 and the (−) face of α10, with the α10 being considered the structural subunit (Plazas et al., 2005). Part of the rationale for this thinking is that because the α9 subunit can assemble into a functional homomer, ACh must bind near the C-loop located on the (+) face of the α9 subunit. Based on these assumptions, (Perez et al. (2009) used molecular modeling to predict binding interactions of α-CTx RgIA with the α9α10 nAChR. The crystal structure of the AChBP bound to α-CTx ImI was used as a template to build these three-dimensional models. MD simulations suggested that α-CTx RgIA interacts with residues on the (+) face of α9 and the (−) face of α10 subunits. This study specifically predicted interaction of α-CTx RgIA R11 and R7 with E194 and with D198 and P197, respectively, in the α9 nAChR subunit, located on the (+) or principal face of the nAChR. The study further predicted interaction of α-CTx RgIA R9 with E58 and D114 in the α10 subunit, located on the (−) or complementary binding site. To comply with the numbering of the α9 nAChR subunit presented in the recent crystal structure of the mature human α9 ECD (Zouridakis et al., 2014), the α9-interacting residues with α-CTx RgIA suggested by Perez et al. (2009) are E197, D201, and P200 in this study, whereas the α10-interacting residues are E61 and D121 (Fig. 1A).
Fig. 1.
Sequence alignments of rat α9 and α10 AChR subunits. (Α) Amino acid sequence alignment of the ECDs of rat α9 and α10 subunits. The α9 and α10 residues studied are indicated by arrows. Alignment was performed with MacVector 10.5.1 ClustalW alignment. (Β) Sequence alignments of the α-CTx RgIA–interacting rat α9 and α10 domains with other nAChR α and β subunits. The residues in the α10(+) and α9(−) faces found to interact with α-CTx RgIA in this study are highlighted. Letters “r” and “h” stand for rat and human, respectively.
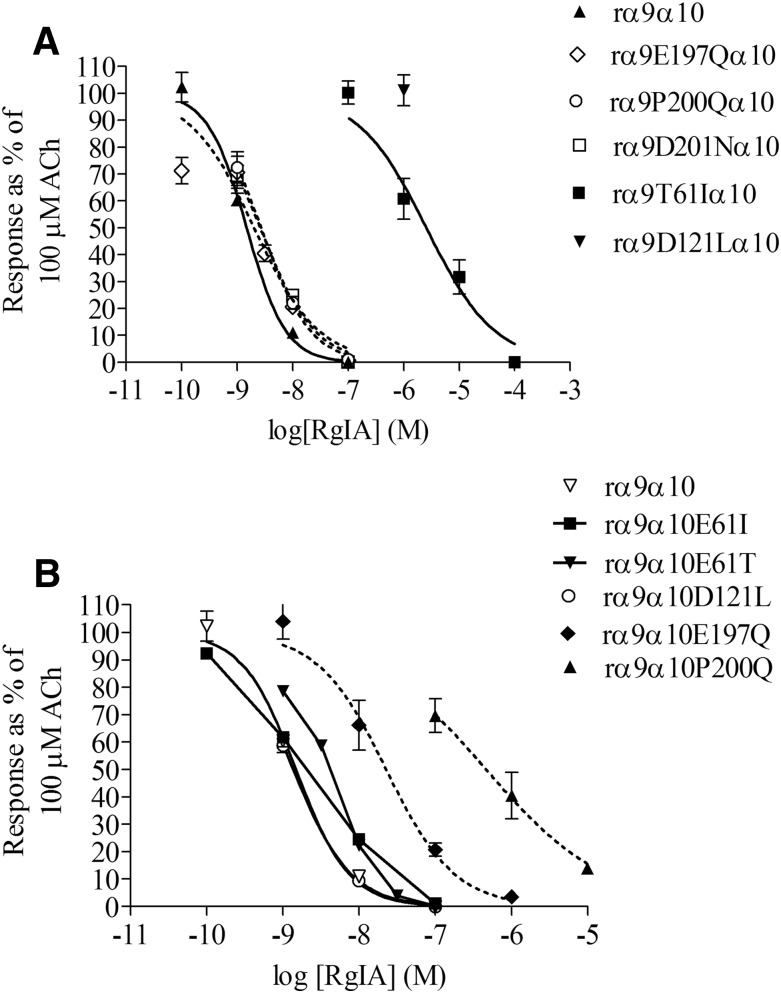
Based on predictions by the Perez et al. (2009) study, we mutated residues in the rat α9(+) and α10(−) binding sites. α9E197Qα10, α9P200Qα10, and α9D201Nα10 were tested for changes in α-CTx RgIA sensitivity. Surprisingly, none of these mutations in the α9(+) binding site had any effect on α-CTx RgIA potency. The potency of α-CTx RgIA on all of these mutants was similar to the wild-type α9α10 nAChR (Fig. 2A; Table 1).
Fig. 2.
Dose-response curves for α-CTx RgIA block of wild-type and mutant α9α10 nAChRs. (A) Mutation of (+) face residues in the α9 subunit did not affect α-CTx potency, whereas mutations in the (−) face residues decreased the potency of α-CTx RgIA. (B) Mutations of the (+) residues of the α10 subunit decreased α-CTx RgIA potency, whereas mutations of the (−) face residues did not. Values are mean ± S.E.M. from at least three different oocytes.
TABLE 1.
IC50 and Hill coefficients for α-CTx RgIA on rat α9 mutant receptors
| nAChR Mutation | IC50 (nM) RgIA (95% CI) | Hill Coefficient |
|---|---|---|
| α9α10 | 1.49 (1.13–1.95) | 1.24 ± 0.19 |
| α9T61Iα10a | 2610 (1230–5330)a | 0.71 ± 0.16 |
| α9T61Eα10a | 28.6 (17.1–47.6)a | 0.51 ± 0.06 |
| α9D121Lα10a | >1000a | ND |
| α9E197Qα10 | 1.15 (0.77–1.73) | 0.61 ± 0.07 |
| α9P200Qα10 | 2.72 (2.13–3.46) | 0.99 ± 0.09 |
| α9D201Nα10 | 2.61 (2.08–3.26) | 0.89 ± 0.07 |
ND, not determined.
Mutations making the greatest difference in toxin potency.
In the (−) face of α10 subunit, E61 was changed to Ile and Thr and D121 to Leu. None of these mutations markedly affected the potency of α-CTx RgIA (Fig. 2B; Table 2). Thus, neither the α9 nor the α10 receptor residues predicted by Perez et al. (2009) to interact with α-CTx RgIA appear to substantially influence binding.
TABLE 2.
IC50 and Hill coefficients for α-CTx RgIA on rat α10 mutant receptors
| nAChR Mutation | IC50 (nM) RgIA (95% CI) | Hill Coefficient |
|---|---|---|
| α9α10 | 1.49 (1.13–1.95) | 1.24 ± 0.19 |
| α9α10E61I | 2.0 (1.24–3.21) | 0.78 ± 0.12 |
| α9α10E61T | 3.54 (2.81–4.47) | 1.17 ± 0.13 |
| α9α10D121L | 1.36 (1.19–1.61) | 1.14 ± 0.09 |
| α9α10E197Qa | 37.5 (26.6–52.8)a | 1.17 ± 0.15 |
| α9α10P200Qa | 466 (176–1230)a | 0.55 ± 0.13 |
Mutations making the greatest difference in toxin potency.
Interestingly, E61 in the α10 subunit, which Perez et al. (2009) suggested to interact with α-CTx RgIA (E58 in their study), corresponds to T61 in the α9 subunit, which in our previous study was shown to contribute to high potency of α-CTx RgIA on the rat versus human α9α10 nAChR (Azam and McIntosh, 2012). This implied that the α-CTx RgIA interaction might occur at an α10/α9 rather than an α9/α10 subunit interface. To test the possibility that the (−) face residues in the α9 and (+) face residues in the α10 subunit interact with α-CTx RgIA [opposite of that suggested by Perez et al. (2009)], we mutated the α10(+) face and the α9(−) face residues at the respective homologous positions of the α9(+) face and α10(−) face.
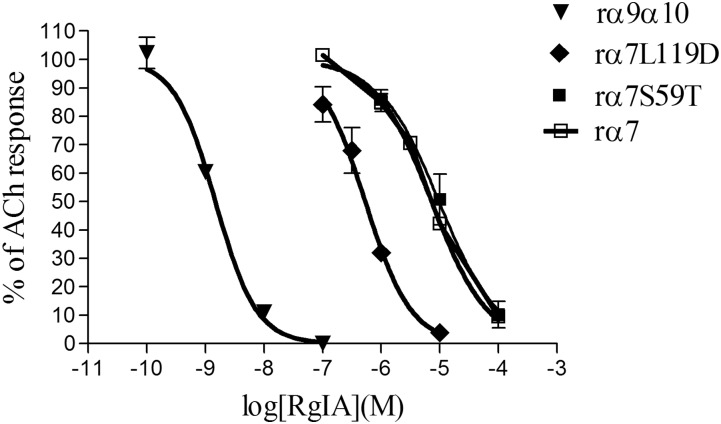
Each mutation of the respective α9(−) face residues caused a loss in α-CTx RgIA potency. More specifically, when α9D121 was mutated to a Leu residue, found in the corresponding region of the α7 subunit, which has low sensitivity to α-CTx RgIA [IC50 of 7.27 μM; 95% confidence interval (CI) 6.13–8.62 μM; Fig. 3], the activity of α-CTx RgIA was nearly abolished on α9D121Lα10; more precisely, no activity was shown at α-CTx RgIA concentrations as high as 1 μM (response after 1 μM α-CTx RgIA: 101 ± 5.7% of control) (Fig. 2A; Table 1;). However, when the converse mutation was made in the α7 subunit, where α7L119 (corresponds to α9D121) was replaced with an Asp, the α7L119D receptor gained more than 10-fold sensitivity to α-CTx RgIA, with an IC50 of 536 nM (95% CI 414–693 nM; Fig. 3). Interestingly, the α7S59T (α7S59 corresponds to rat α9T61) did not change sensitivity toward α-CTx RgIA (Fig. 3), suggesting that Thr in this position is not imperative for α-CTx RgIA binding to all subunits. Concerning the rat α9T61 residue, because we had previously shown that its replacement with an Ile residue, found in the homologous position in the human α9 subunit, dramatically affected α-CTx RgIA potency (Azam and McIntosh, 2012) (Table 1), we now sought to determine whether mutation of this Thr residue to something other than an Ile still had an effect on α-CTx RgIA activity. We chose Glu, as this residue is found in the homologous position in the α10 subunit and was predicted to interact with R9 of α-CTx RgIA (Perez et al., 2009). α9T61E had an almost 20-fold lower sensitivity to α-CTx RgIA compared with wild-type receptor (Table 1).
Fig. 3.
Dose-response curves for α-CTx RgIA on rat α9α10, rat α7, and rat α7 mutants α7S59T and α7L119D. There was an approximately 10-fold gain in potency of RgIA on the α7L119D receptor mutant. The concentrations of ACh used were 100 μM for α9α10 and 200 μM for α7 and α7 mutants. Data are mean ± S.E.M. from at least three separate oocytes.
Likewise, when the (+) face residues of the α10 subunit were mutated, the potency of α-CTx RgIA was affected. α9α10E197Q and α9α10P200Q were 25- and 300-fold, respectively, less sensitive to α-CTx RgIA compared with wild-type α9α10 nAChR (Fig. 2B; Table 2). Unfortunately, the α10D201N mutation did not consistently yield functional expression and therefore was not tested.
Previous studies in Xenopus oocytes have indicated that Ca2+ entry (such as through opening of α9α10 nAChRs) can lead to activation of Ca2+-dependent Cl− channels (Barish, 1983). At the holding potential used in this study (−70 mV), this can cause a large negative current, in addition to the inward positive current through the α9α10 nAChRs. Therefore, we performed a set of electrophysiological experiments using a buffer in which Ba2+ was used in place of Ca2+ to exclude the possibility that block by the toxin was mediated by block of Ca2+-dependent Cl− channels rather than block of α9α10 nAChRs. In Ba2+ buffer, measured currents were ≤10% of those observed in the Ca2+ buffer (L. Azam, unpublished data). However, the potency of α-CTx RgIA for blocking the α9α10 nAChR in the Ba2+ (2.05 nM; 95% CI 1.48–2.85 nM) did not significantly differ from that observed in regular ND-96 buffer (1.49 nM; 95% CI 1.13–1.95 nM). Unfortunately, when most of the receptors formed by the mutant α9 or α10 subunits were tested in the Ba2+ buffer, the currents obtained were too small for accurate measurement of α-CTx RgIA potency. Only one of the mutants, α9T61Iα10, gave robust currents in the Ba2+-containing buffer. The IC50 for block of α-CTx RgIA was similar in Ca2+-containing (2.61 μM; Table 1) and Ca2+-free (4.93 μM, 95% CI 2.4–11.4 μM) buffers.
Effect of Mutations in the α9 and α10 Subunits Affecting RgIA on ACh Sensitivity
The mutations in both the α9 and the α10 subunits that affected α-CTx RgIA potency were tested for any changes in ACh sensitivity. ACh activated wild-type rat α9α10 nAChR with an EC50 value of 10.1 μM (Table 3). The mutated (+) face α10 residues E197 and P200 caused an approximately 20-fold (mutant α10E197Q) and a 40-fold rightward shift (mutant α10P200Q) in ACh potency, respectively (Fig. 4; Table 3). The mutated (−) face α9 residues had differential effects on ACh potency. The α9T61I mutant that caused a >1500-fold change in α-CTx RgIA potency (see above) did not affect ACh potency. However, α9D121Lα10 lowered the ACh potency by about 30-fold (Fig. 4; Table 3;).
TABLE 3.
EC50 for ACh dose-response curves on α9 and α10 mutants that affected α-CTx RgIA potency
Values are mean ± S.E.M. from at least three different oocytes.
| nAChR | ACh EC50 (95% CI) μM | Hill Slope |
|---|---|---|
| α9α10 | 10.1 (8.68–11.8) | 1.2 ± 0.1 |
| α9T61Iα10 | 10.7 (8.90–12.9) | 1.4 ± 0.2 |
| α9D121Lα10 | 323 (238–437) | 1.8 ± 0.4 |
| α9α10E197Q | 193 (145–257) | 0.54 ± 0.04 |
| α9α10P200Q | 419 (369–477) | 0.81 ± 0.04 |
Fig. 4.
Agonist dose-response curves for α9 and α10 mutant receptors. Responses to a brief pulse of ACh on α9α10 nAChR mutants were measured as described in Materials and Methods. Data are mean ± S.E.M. from at least three separate oocytes.
Molecular Modeling of the Rat α9α10 ECD Based on the X-ray Crystal Structure of the Human α9 ECD
Molecular Modeling Studies of the Complex of α9α10 ECD with α-CTx RgIA.
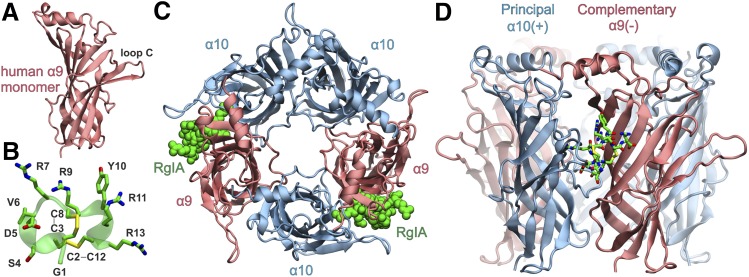
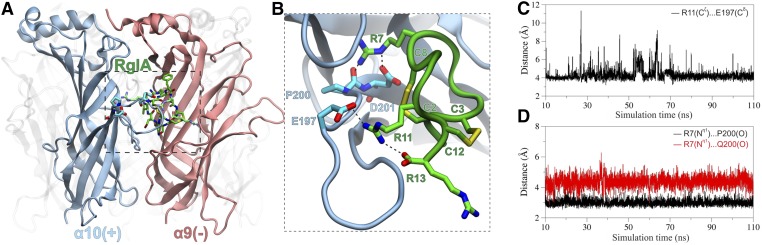
To gain insight into the molecular basis of the interaction between the rat nAChR and α-CTx RgIA, we employed MD calculations of the ECD of the rat (α9)2(α10)3 nAChR complex with α-CTx RgIA. Our model was based on the recent X-ray crystal structure of the monomeric state of the ECD of the human α9 nAChR (Fig. 5A) (Zouridakis et al., 2014) and the solution NMR structure of α-CTx RgIA (Fig. 5B) (Ellison et al., 2008). Molecular models of the highly homologous rat α9 and α10 subunits were assembled in the pentameric state (Fig. 5, C and D) on the basis of the AChBP complex with α-CTx ImI, as described in Materials and Methods. Two α-CTx molecules were modeled in the two α10(+)/α9(−) binding sites (Fig. 5C), and unrestrained MD simulations in explicit solvent were carried out (Supplement Material 1). The systems exhibited significant stability during the course of the MD simulations as demonstrated by the root-mean-square deviations from the initial conformation and the radius of gyration of the receptor (Supplemental Figs. 2 and 3). The bound α-CTx molecules displayed similar arrangement within the two α10/α9 sites, but did not exhibit identical interactions as a function of simulation time, especially with respect to the C-terminal R13 (see below).
Fig. 5.
(A) Ribbon representation of the X-ray crystal structure of the monomeric human α9 ECD (PDB ID 4UXU) employed as template for the homology modeling of the rat α9 and α10 subunits. (B) Representative structure from the solution NMR conformational ensemble of α-CTx RgIA (PDB ID 2JUQ). The side chains are shown in stick representation with C in green, N in blue, O in red, and S in yellow. (C) Molecular model of the ECD of the rat (α9)2(α10)3 complex with α-CTx RgIA (green spheres). The arrangement of subunits is based on the X-ray crystal structure of Aplysia californica AChBP in complex with α-CTx ImI (PDB ID 2BYP). α-CTx RgIA was superimposed with α-CTx ImI at the two α10/α9 ligand binding sites. (D) Side view of the α10/α9 binding site with bound α-CTx RgIA (stick representation), where α10(+) is designated as the principal subunit and α9(−) as the complementary subunit.
At the α10(+) side (Fig. 6A), E197 displayed a quite stable electrostatic interaction with the side chain of R11 (Fig. 6, B and C). Therefore, the significant decrease in the sensitivity to α-CTx RgIA for the mutant α9α10E197Q (Table 2) could be due to the impairment of the formation of a salt bridge with R11. In addition, the carbonyl group of P200 formed a stable hydrogen bond with the guanidinium group of R7, which was stabilized by an electrostatic interaction between D201 and R7 (Fig. 6B). Because the predicted interaction of R7 with the main chain of P200 cannot readily explain the observed reduction in the potency of α-CTx RgIA by 300 times for the α9α10P200Q mutant, we carried out MD of the same model, but with a single P200Q mutation at one of the two α10α9 sites. In this case, our calculations revealed that the guanidinium group of R7 and the backbone C = O group of Q200 were not within hydrogen-bonding distance throughout the course of the MD simulations (Fig. 6D). This can be justified by the higher flexibility of Gln with respect to Pro, which allows a rotation of the backbone carbonyl group away from R7. Although we were not able to obtain functional expression for the α10D201N mutation, our modeling data indicated that the interaction between R7 and α10D201 was preserved during the MD simulations (Supplemental Fig. 4), which suggests the importance of an Asp residue in this position for binding of α-CTx RgIA.
Fig. 6.
(A) Representative structure of the top-populated conformational cluster from the MD simulations of the rat (α9)2(α10)3 complex with α-CTx RgIA indicating the position of the three α10(+) residues E197, P200, and D201. (B) Close-up view of the α10/α9–binding interface illustrating the interactions between R7 and R11 of α-CTx RgIA, and the α10(+) residues E197, P200, and D201. Residues from the α10 subunit are shown with carbon atoms in cyan, and all other colors are as in Fig. 5. (C) Plot of the distance between R11-Cζ and E197-Cδ during the course of the MD production runs. (D) Plot of the distance between the R7- Nη1 atom and P200-O (black line) or Q200-O (red line) in the α9α10P200Q mutant.
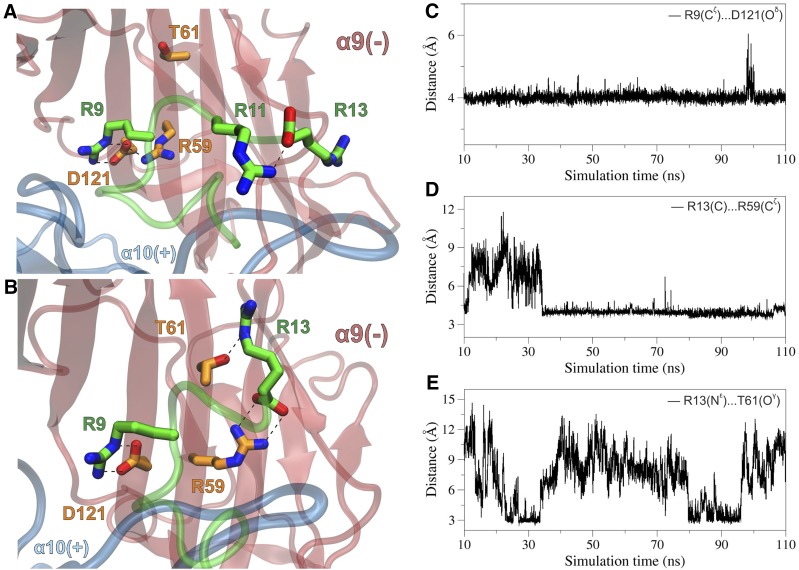
At the α9(−) face, D121 formed a stable salt bridge interaction with R9 of α-CTx RgIA throughout the MD simulations in both α10α9 sites (Fig. 7, A–C), whose importance was depicted at the >670-fold decrease in the potency of α-CTx RgIA in the α9D121Lα10 mutant (Table 1). In one α10/α9 site, D121 also exhibited a stable hydrogen-bonding intramolecular interaction with R59, which was also apparent at the X-ray crystal structure of the monomeric human α9 ECD (Zouridakis et al., 2014). At this site, our simulations did not reveal any direct interaction between the antagonist and T61, rather than water-mediated interactions between the α9 T61 hydroxyl group and the backbone of the R9-Y10-R11 moiety. At the other α10/α9 binding site, R59 shifted away from D121 and formed a salt bridge interaction with R13 of α-CTx RgIA (Fig. 7, B and D). This movement had no effect in the intermolecular interaction between α9 D121 and R9 of α-CTx RgIA; however, the side chain of R13 displayed hydrogen-bonding interactions with the hydroxyl group of α9 T61, albeit for a fraction of the simulation time as a result of the high mobility of R13 (Fig. 7, B and E). Therefore, the significant effect of the α9T61Iα10 mutant in the potency of α-CTx RgIA (Fig. 2; Table 3) could be attributed to the loss of a potent hydrogen bond with R13 of α-CTx RgIA and/or the altered hydration degree of the α9(−) site.
Fig. 7.
(A) Close-up view of the α-CTx–binding interface from a representative structure of the rat (α9)2(α10)3 ECD complex with α-CTx RgIA, illustrating the interactions between R9 of α-CTx RgIA and D121, which forms a stable salt bridge interaction with R59 at the α9(−) subunit. Residues from α9 subunit are shown with carbon atoms in orange, and all other colors are as in Fig. 5. (B) Close-up view of the second α10/α9–binding interface from a snapshot of the MDs of the rat (α9)2(α10)3 complex with α-CTx RgIA, illustrating the potential hydrogen-bonding interactions of R13 of α-CTx RgIA with R59 and T61 at the α9(−) subunit. (C) Plot of the distance between R9-Cζ and D121-Oδ from the first α10/α9 site as function of simulation time. (D) Plot of the distance between the C-terminus R13-C and R59-Cζ from the second α10/α9 site as a function of simulation time. (E) For the same site, plot of the distance between R13-Nε and T61-Oγ during the course of the MDs.
Nicotinic receptors are members of the Cys-loop ligand-gated ion channel superfamily. Other evolutionarily related members include GABAA, glycine, and 5HT3 receptors. Each of these is formed from five subunits arranged around a central ion-conducting pore; each subunit has a characteristic Cys-loop formed by two highly conserved Cys residues of the ECD. We aligned the ECDs of these receptors to examine possible homology with the critical residues identified for the α9 and α10 nAChRs. E197 was conserved in the 5HT3 receptor, but the other residues were not conserved among the non-nicotinic receptors (Supplemental Fig. 5). This is consistent with α-CTx RgIA having an IC50 > 10 µM for the 5-HT3 and GABAA receptor in competitive binding assays (unpublished results), in contrast to potent block of α9α10 nAChRs.
Molecular Modeling Studies of the Complex of α9α10 ECD with ACh.
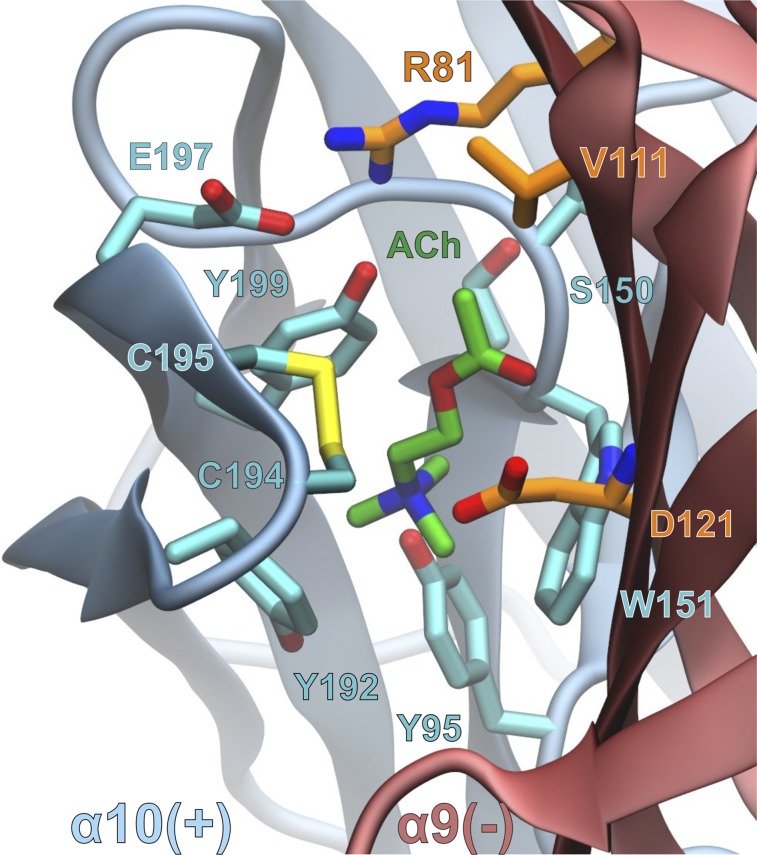
In the molecular model of the rat (α9)2(α10)3 nAChR complex with ACh, the agonist is surrounded by an aromatic cage comprising Y95, Y192, Y199, and W151 from the α10(+) subunit and displays additional interactions with the C194–C195 disulfide bridge, S150 and E197 (Fig. 8; Supplemental Material 2). Interestingly, α10E197 formed a salt bridge with α9R81; therefore, it is possible that the α10E197Q mutation disrupted the optimum conformation of the α10/α9 binding site, as demonstrated by the 20-fold increase in the EC50 value for ACh (Table 3). Similarly, it is possible that the α10P200Q mutation resulted in local conformational rearrangements that affected the interaction of ACh with the neighboring Y199 residue by 40-fold, respectively (Table 3). At the α9(−) side, the carboxylate group of D121 is predicted to form electrostatic interactions with the trimethyl ammonium moiety of ACh, the impairment of which led to a 30-fold increase of the EC50 value in the α9D121Lα10 mutant (Table 3). In addition, V111 and the aliphatic moiety of R81 formed van der Waals interactions with the acetoxy group of ACh (Fig. 8).
Fig. 8.
Close-up view of the α10/α9–binding interface from a representative structure of the MD simulations of rat (α9)2(α10)3 complex with ACh. The interacting residues within 4.5 Å from the ligand (green carbon atoms) are shown with cyan and orange carbon atoms for α10(+) and α9(−) subunits, respectively.
Discussion
In this study, we demonstrated asymmetric interaction of α-CTx RgIA with the rat α9α10 nAChR. The mutational studies indicated interaction of α-CTx RgIA with the α10(+)/α9(−) interface of the α9α10 nAChR, which is opposite to that proposed by previous modeling studies based on the structure of the homologous AChBP from invertebrates (Perez et al., 2009). Interestingly, however, the residues of interaction of the α10(+) and of the α9(−) faces found in our study are in homologous positions to those predicted by Perez et al. (2009) for the opposite subunits. Thus, the Perez et al. (2009) model, although incorrect in assignment of principal and complementary binding site subunits, succeeded in assigning interacting residues if subunit interface is not considered. Based on the recent X-ray crystal structure of the ECD of human α9 nAChR, we performed MD simulations of the rat α9α10 ECD complexes with α-CTx RgIA to provide a molecular basis for the results of our functional studies. The current model is probably more accurate than the previous ones based on the homologous AChBPs, because the sequence identities of rat α9 and α10 ECDs with human α9 ECD are 96 and 67%, respectively, whereas the identities with AChBP are below 25%. Indeed, the results of these simulations were consistent with our functional studies.
The muscle nAChR was the first nicotinic receptor to be rigorously examined with respect to selective toxin binding to subunit interface. Two ACh binding sites are formed between its α1/δ and α1/γ (or ε in adult) interfaces. Sine et al. (1995) demonstrated that a paralytic peptide, α-CTx MI, selectively binds the α1/δ interface (Sine et al., 1995; Sugiyama et al., 1998); subsequently, other toxins were discovered that selectively bind the α1/δ, α1/ε, or α1/γ interfaces (Hann et al., 1994; Groebe et al., 1995; Martinez et al., 1995; Molles et al., 2002a,b), which allowed the development of radiolabeled and fluorescent probes for specific labeling of fetal or adult muscle nAChR subtypes (Teichert et al., 2008).
Neuronal nAChRs are composed of α and β subunits. Their ligand binding sites have been shown to be formed at the interfaces between the (+) face of an α subunit and the (−) face of a β subunit; several conotoxins have been discovered that discriminate between the different subunit interfaces. α-CTx MII (Cartier et al., 1996), for example, shows high discrimination between α6/β2 and α4/β2 subunit interfaces (Salminen et al., 2004). Thus, α-CTxs have greatly contributed to the molecular dissection of neuronal nAChR subtypes that modulate the release of neurotransmitters. The present study shows that α-CTx RgIA can discriminate among asymmetric heteromeric α/α subunit interfaces; in the rat α9α10 nAChR, α-CTx RgIA selectively interacts with α10(+)/α9(−) versus α9(+)/α10(−) interfaces. The α9 subunit, in contrast to the α10, forms homopentamers, and that is one reason it was originally assumed that competitive ligands would bind to the (+) face of the α9 subunit and (−) face of either α9 (homomeric nAChR) or (−) face of α10 in the heteromeric α9α10 nAChR. In support of this, Ellison et al. (2008) showed that mutation of W176 (W151 based on numbering in this study) to Thr in the α9(+) face resulted in loss of potency for ACh and α-CTx RgIA, consistent with competitive interaction of α-CTx RgIA at an agonist binding site. In addition, crystallization of the ECD of the human α9 subunit in the presence of antagonists indicated a major contribution of the α9(+) face for binding of methyllycaconitine and α-bungarotoxin (Zouridakis et al., 2014). However, the present study showed that there is a separate, high-affinity, binding site for α-CTx RgIA and ACh, which is the α10(+)/α9(−) interface.
Based on the model of rat α9α10 nAChR suggested by Perez et al. (2009), we mutated the α9(+) and α10(−) face residues shown to interact with α-CTx RgIA and investigated their effect. The α9(+) face residues proposed to interact with R11 and R7 of the toxin were E194 and P197/D198, respectively. When any of these residues was mutated in the α9 subunit (E197, P200, and D201 in this study), there was no effect on α-CTx RgIA potency (Table 1). In contrast, when the same residues in the α10(+) face were mutated (α10 Ε197 and P200Q), there was a 25- to 300-fold loss in toxin activity (Table 2). This effect is much larger than the 5-fold reduction in potency for the W176T mutation in the α9(+) face found in the Ellison et al. (2008) study (W151T based on our numbering). In addition, both the E197Q and P200Q mutations in the α10(+) face that affect α-CTx RgIA binding also decrease the potency of ACh for activation of the α9α10 nAChR (Table 3).
Our molecular modeling studies suggested putative interactions between α10Ε197 and R11 of α-CTx RgIA, as well as between α10P200, α10D201, and R7 of α-CTx RgIA in the α10(+)/α9(−)–binding interface (Fig. 6), in support to our mutational studies. Interestingly, in all other α subunits, conferring the (+) faces of ACh binding sites, with the exception of α5 (which has not been reported to participate in ACh binding sites), a negatively charged residue exists in the homologous positions to α10 Ε197 and D201 (Fig. 1B). P200, whose main chain is predicted to interact with α-CTx RgIA, is also highly conserved, with the exception of α1 and α6, in which a Tyr or a Thr residue exists at this position, respectively (Fig. 1B). This Thr in the α6 subunit has been shown to confer high potency of an α-CTx MII analog on the α6β2β3 nAChR (Azam et al., 2008).
In addition, Perez et al. (2009) also suggested a role for the α10(−) face residues E61 and D121 (our numbering) in interacting with α-CTx RgIA. However, we did not observe a dramatic change in α-CTx RgIA potency when either residue in the α10 subunit was mutated to an Ile/Thr or Leu, respectively (Table 2). Instead, D121L mutation in the α9(−) face caused a complete loss of sensitivity to α-CTx RgIA (Table 1). We chose Leu as a substitute for Asp, because this residue exists in the homologous position of the α7 subunit (α7 L119), which has low sensitivity to α-CTx RgIA (Ellison et al., 2006). Consistent with this, when the converse mutation was made in the α7 subunit, the homopentamer formed from the mutant subunit α7L119D had >10-fold higher sensitivity to α-CTx RgIA. According to our model, D121 of the α9(−) face forms a salt bridge with R9 of α-CTx RgIA, similar to the D121 of the α10(−) face in the Perez et al. (2009) study. Notably, among nAChR subunits, this negatively charged residue is only found in the α9 and α10 subunits (Fig. 1B), suggesting that α9 D121 is critical for activity and selectivity of α-CTx RgIA for the α9α10 nAChR.
In position 61 of the rat α9 subunit, a Thr residue exists instead of Glu, which is present in α10 (Fig. 1A). A Glu residue in this position also exists in the (−) face of the binding sites of β2 and β4 subunits. α9T61 was previously shown (Azam and McIntosh, 2012) to confer ∼300-fold higher potency of α-CTx RgIA on the rat versus human α9 subunit, which instead has an Ile residue at this position (Fig. 1B). In addition, when we mutated α9T61 to Glu (found in α10, β2, β3, and β4) in the present study, a ∼20-fold decrease in the potency for α-CTx RgIA was observed (Table 1), confirming that α9 confers the (−) face of α-CTx binding site in the α9α10 nAChR. Our MD simulations of the rat α10(+)/α9(−)–binding interface displayed water-mediated interactions of α9T61 with the R9-Y10-R11 moiety of α-CTx RgIA, as well as the potential formation of a hydrogen bond with R13 (Fig. 7). The importance of a Thr residue at the (−) face of an α-CTx–interacting nAChR subunit has precedents. In the β2 subunit, T59, which is two residues away from the homologous position to α9T61 (Fig. 1B), is a determinant of selectivity for α-CTx MII on the α3β2 nAChR (Harvey et al., 1997), and a critical residue in off-rate kinetics of α-CTx BuIA on the β2 subunit (Shiembob et al., 2006).
Taken together, our results strongly support the existence of an additional binding site for ACh in the α9α10 nAChR between the α10(+)/α9(−) interface (Fig. 8). This α10(+)/α9(−) interface is the high-affinity binding site for α-CTx RgIA binding and consistent with a recent study, which also suggested a similar interaction for α-CTx Vc1.1 with the α10(+)/α9(−) interface of the rat α9α10 nAChR (Yu et al., 2013). Conotoxins that bind the α9α10 nAChR are being considered as potential pain therapeutics (McIntosh et al., 2009; Del Bufalo et al., 2014; Di Cesare Mannelli et al., 2014), and understanding the subunit determinants of α-CTx interaction with the α9α10 nAChR may facilitate further development of such compounds.
Supplementary Material
Abbreviations
- ACh
acetylcholine
- AChBP
ACh-binding protein
- α-CTx
α-conotoxin
- CI
confidence interval
- ECD
extracellular domain
- MD
molecular dynamics
- nAChR
nicotinic ACh receptor
Authorship Contributions
Participated in research design: Azam, McIntosh.
Conducted experiments: Azam, Papakyriakou.
Contributed new reagents or analytic tools: Azam, Papakyriakou, Zouridakis, Giastas, Tzartos, McIntosh.
Performed data analysis: Azam, Papakyriakou, Giastas, Zouridakis, Tzartos.
Wrote or contributed to the writing of the manuscript: Azam, Papakyriakou, Zouridakis, Giastas, Tzartos, McIntosh.
Footnotes
This work was supported by National Institutes of Health [Grants P01-GM48677, R01-GM103801] and European Commission Seventh Framework Programme ‘REGPOT-NeuroSign’ [Grant 264083].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Albuquerque EX, Pereira EF, Alkondon M, Rogers SW. (2009) Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev 89:73–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam L, McIntosh JM. (2012) Molecular basis for the differential sensitivity of rat and human α9α10 nAChRs to α-conotoxin RgIA. J Neurochem 122:1137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam L, Yoshikami D, McIntosh JM. (2008) Amino acid residues that confer high selectivity of the alpha6 nicotinic acetylcholine receptor subunit to alpha-conotoxin MII[S4A,E11A,L15A]. J Biol Chem 283:11625–11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish ME. (1983) A transient calcium-dependent chloride current in the immature Xenopus oocyte. J Physiol 342:309–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzat C. (2012) New insights into the structural bases of activation of Cys-loop receptors. J Physiol 106:23–33. [DOI] [PubMed] [Google Scholar]
- Cartier GE, Yoshikami D, Gray WR, Luo S, Olivera BM, McIntosh JM. (1996) A new alpha-conotoxin which targets alpha3beta2 nicotinic acetylcholine receptors. J Biol Chem 271:7522–7528. [DOI] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celie PH, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. (2004) Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 41:907–914. [DOI] [PubMed] [Google Scholar]
- Colomer C, Olivos-Oré LA, Vincent A, McIntosh JM, Artalejo AR, Guérineau NC. (2010) Functional characterization of alpha9-containing cholinergic nicotinic receptors in the rat adrenal medulla: implication in stress-induced functional plasticity. J Neurosci 30:6732–6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bufalo A, Cesario A, Salinaro G, Fini M, Russo P. (2014) Alpha9 alpha10 nicotinic acetylcholine receptors as target for the treatment of chronic pain. Curr Pharm Des 20:6042–6047. [DOI] [PubMed] [Google Scholar]
- Di Cesare Mannelli L, Cinci L, Micheli L, Zanardelli M, Pacini A, McIntosh JM, Ghelardini C. (2014) Α-conotoxin RgIA protects against the development of nerve injury-induced chronic pain and prevents both neuronal and glial derangement. Pain 155:1986–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgoyhen AB, Johnson DS, Boulter J, Vetter DE, Heinemann S. (1994) Alpha 9: an acetylcholine receptor with novel pharmacological properties expressed in rat cochlear hair cells. Cell 79:705–715. [DOI] [PubMed] [Google Scholar]
- Elgoyhen AB, Vetter DE, Katz E, Rothlin CV, Heinemann SF, Boulter J. (2001) alpha10: a determinant of nicotinic cholinergic receptor function in mammalian vestibular and cochlear mechanosensory hair cells. Proc Natl Acad Sci USA 98:3501–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison M, Feng ZP, Park AJ, Zhang X, Olivera BM, McIntosh JM, Norton RS. (2008) Alpha-RgIA, a novel conotoxin that blocks the alpha9alpha10 nAChR: structure and identification of key receptor-binding residues. J Mol Biol 377:1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison M, Haberlandt C, Gomez-Casati ME, Watkins M, Elgoyhen AB, McIntosh JM, Olivera BM. (2006) Alpha-RgIA: a novel conotoxin that specifically and potently blocks the alpha9alpha10 nAChR. Biochemistry 45:1511–1517. [DOI] [PubMed] [Google Scholar]
- Fiser A, Sali A. (2003) Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 374:461–491. [DOI] [PubMed] [Google Scholar]
- Gotti C, Moretti M, Gaimarri A, Zanardi A, Clementi F, Zoli M. (2007) Heterogeneity and complexity of native brain nicotinic receptors. Biochem Pharmacol 74:1102–1111. [DOI] [PubMed] [Google Scholar]
- Gotti C, Zoli M, Clementi F. (2006) Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol Sci 27:482–491. [DOI] [PubMed] [Google Scholar]
- Groebe DR, Dumm JM, Levitan ES, Abramson SN. (1995) alpha-Conotoxins selectively inhibit one of the two acetylcholine binding sites of nicotinic receptors. Mol Pharmacol 48:105–111. [PubMed] [Google Scholar]
- Haberberger RV, Bernardini N, Kress M, Hartmann P, Lips KS, Kummer W. (2004) Nicotinic acetylcholine receptor subtypes in nociceptive dorsal root ganglion neurons of the adult rat. Auton Neurosci 113:32–42. [DOI] [PubMed] [Google Scholar]
- Hann RM, Pagán OR, Eterović VA. (1994) The alpha-conotoxins GI and MI distinguish between the nicotinic acetylcholine receptor agonist sites while SI does not. Biochemistry 33:14058–14063. [DOI] [PubMed] [Google Scholar]
- Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, Bourne Y. (2005) Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J 24:3635–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SC, McIntosh JM, Cartier GE, Maddox FN, Luetje CW. (1997) Determinants of specificity for alpha-conotoxin MII on alpha3beta2 neuronal nicotinic receptors. Mol Pharmacol 51:336–342. [DOI] [PubMed] [Google Scholar]
- Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65:712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. (1996) VMD: visual molecular dynamics. J Mol Graph 14:33-38, 27-28. [DOI] [PubMed] [Google Scholar]
- Katz E, Elgoyhen AB, Gómez-Casati ME, Knipper M, Vetter DE, Fuchs PA, Glowatzki E. (2004) Developmental regulation of nicotinic synapses on cochlear inner hair cells. J Neurosci 24:7814–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Meizel S. (2005) Nicotinic acetylcholine receptor subunits and associated proteins in human sperm. J Biol Chem 280:25928–25935. [DOI] [PubMed] [Google Scholar]
- Kurzen H, Berger H, Jäger C, Hartschuh W, Näher H, Gratchev A, Goerdt S, Deichmann M. (2004) Phenotypical and molecular profiling of the extraneuronal cholinergic system of the skin. J Invest Dermatol 123:937–949. [DOI] [PubMed] [Google Scholar]
- Lester HA, Dibas MI, Dahan DS, Leite JF, Dougherty DA. (2004) Cys-loop receptors: new twists and turns. Trends Neurosci 27:329–336. [DOI] [PubMed] [Google Scholar]
- Lips KS, Pfeil U, Kummer W. (2002) Coexpression of alpha 9 and alpha 10 nicotinic acetylcholine receptors in rat dorsal root ganglion neurons. Neuroscience 115:1–5. [DOI] [PubMed] [Google Scholar]
- Lustig LR. (2006) Nicotinic acetylcholine receptor structure and function in the efferent auditory system. Anat Rec A Discov Mol Cell Evol Biol 288:424–434. [DOI] [PubMed] [Google Scholar]
- Lustig LR, Peng H, Hiel H, Yamamoto T, Fuchs PA. (2001) Molecular cloning and mapping of the human nicotinic acetylcholine receptor alpha10 (CHRNA10). Genomics 73:272–283. [DOI] [PubMed] [Google Scholar]
- Martinez JS, Olivera BM, Gray WR, Craig AG, Groebe DR, Abramson SN, McIntosh JM. (1995) alpha-Conotoxin EI, a new nicotinic acetylcholine receptor antagonist with novel selectivity. Biochemistry 34:14519–14526. [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Absalom N, Chebib M, Elgoyhen AB, Vincler M. (2009) Alpha9 nicotinic acetylcholine receptors and the treatment of pain. Biochem Pharmacol 78:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JM, Plazas PV, Watkins M, Gomez-Casati ME, Olivera BM, Elgoyhen AB. (2005) A novel alpha-conotoxin, PeIA, cloned from Conus pergrandis, discriminates between rat alpha9alpha10 and alpha7 nicotinic cholinergic receptors. J Biol Chem 280:30107–30112. [DOI] [PubMed] [Google Scholar]
- Millar NS, Gotti C. (2009) Diversity of vertebrate nicotinic acetylcholine receptors. Neuropharmacology 56:237–246. [DOI] [PubMed] [Google Scholar]
- Molles BE, Rezai P, Kline EF, McArdle JJ, Sine SM, Taylor P. (2002a) Identification of residues at the alpha and epsilon subunit interfaces mediating species selectivity of Waglerin-1 for nicotinic acetylcholine receptors. J Biol Chem 277:5433–5440. [DOI] [PubMed] [Google Scholar]
- Molles BE, Tsigelny I, Nguyen PD, Gao SX, Sine SM, Taylor P. (2002b) Residues in the epsilon subunit of the nicotinic acetylcholine receptor interact to confer selectivity of waglerin-1 for the alpha-epsilon subunit interface site. Biochemistry 41:7895–7906. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Ndoye A, Grando SA. (2000) Novel human alpha9 acetylcholine receptor regulating keratinocyte adhesion is targeted by Pemphigus vulgaris autoimmunity. Am J Pathol 157:1377–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Ferris RL, Matthews T, Hiel H, Lopez-Albaitero A, Lustig LR. (2004) Characterization of the human nicotinic acetylcholine receptor subunit alpha (alpha) 9 (CHRNA9) and alpha (alpha) 10 (CHRNA10) in lymphocytes. Life Sci 76:263–280. [DOI] [PubMed] [Google Scholar]
- Pérez EG, Cassels BK, Zapata-Torres G. (2009) Molecular modeling of the alpha9alpha10 nicotinic acetylcholine receptor subtype. Bioorg Med Chem Lett 19:251–254. [DOI] [PubMed] [Google Scholar]
- Plazas PV, Katz E, Gomez-Casati ME, Bouzat C, Elgoyhen AB. (2005) Stoichiometry of the alpha9alpha10 nicotinic cholinergic receptor. J Neurosci 25:10905–10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux I, Wersinger E, McIntosh JM, Fuchs PA, Glowatzki E. (2011) Onset of cholinergic efferent synaptic function in sensory hair cells of the rat cochlea. J Neurosci 31:15092–15101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen O, Murphy KL, McIntosh JM, Drago J, Marks MJ, Collins AC, Grady SR. (2004) Subunit composition and pharmacology of two classes of striatal presynaptic nicotinic acetylcholine receptors mediating dopamine release in mice. Mol Pharmacol 65:1526–1535. [DOI] [PubMed] [Google Scholar]
- Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC. (2013) Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald J Chem Theory Comput 9:3878–3888. [DOI] [PubMed] [Google Scholar]
- Sgard F, Charpantier E, Bertrand S, Walker N, Caput D, Graham D, Bertrand D, Besnard F. (2002) A novel human nicotinic receptor subunit, alpha10, that confers functionality to the alpha9-subunit. Mol Pharmacol 61:150–159. [DOI] [PubMed] [Google Scholar]
- Shiembob DL, Roberts RL, Luetje CW, McIntosh JM. (2006) Determinants of alpha-conotoxin BuIA selectivity on the nicotinic acetylcholine receptor beta subunit. Biochemistry 45:11200–11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard AR, Gan Y, St-Pierre S, Kousari A, Patel V, Whiteaker P, Morley BJ, Lukas RJ, Shi FD. (2013) Differential modulation of EAE by α9*- and β2*-nicotinic acetylcholine receptors. Immunol Cell Biol 91:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sine SM, Engel AG. (2006) Recent advances in Cys-loop receptor structure and function. Nature 440:448–455. [DOI] [PubMed] [Google Scholar]
- Sine SM, Kreienkamp HJ, Bren N, Maeda R, Taylor P. (1995) Molecular dissection of subunit interfaces in the acetylcholine receptor: identification of determinants of alpha-conotoxin M1 selectivity. Neuron 15:205–211. [DOI] [PubMed] [Google Scholar]
- Sugiyama N, Marchot P, Kawanishi C, Osaka H, Molles B, Sine SM, Taylor P. (1998) Residues at the subunit interfaces of the nicotinic acetylcholine receptor that contribute to alpha-conotoxin M1 binding. Mol Pharmacol 53:787–794. [DOI] [PubMed] [Google Scholar]
- Teichert RW, Garcia CC, Potian JG, Schmidt JJ, Witzemann V, Olivera BM, McArdle JJ. (2008) Peptide-toxin tools for probing the expression and function of fetal and adult subtypes of the nicotinic acetylcholine receptor. Ann N Y Acad Sci 1132:61–70. [DOI] [PubMed] [Google Scholar]
- Vetter DE, Katz E, Maison SF, Taranda J, Turcan S, Ballestero J, Liberman MC, Elgoyhen AB, Boulter J. (2007) The alpha10 nicotinic acetylcholine receptor subunit is required for normal synaptic function and integrity of the olivocochlear system. Proc Natl Acad Sci USA 104:20594–20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter DE, Liberman MC, Mann J, Barhanin J, Boulter J, Brown MC, Saffiote-Kolman J, Heinemann SF, Elgoyhen AB. (1999) Role of alpha9 nicotinic ACh receptor subunits in the development and function of cochlear efferent innervation. Neuron 23:93–103. [DOI] [PubMed] [Google Scholar]
- Vincler M, Wittenauer S, Parker R, Ellison M, Olivera BM, McIntosh JM. (2006) Molecular mechanism for analgesia involving specific antagonism of alpha9alpha10 nicotinic acetylcholine receptors. Proc Natl Acad Sci USA 103:17880–17884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174. [DOI] [PubMed] [Google Scholar]
- Yu R, Kompella SN, Adams DJ, Craik DJ, Kaas Q. (2013) Determination of the α-conotoxin Vc1.1 binding site on the α9α10 nicotinic acetylcholine receptor. J Med Chem 56:3557–3567. [DOI] [PubMed] [Google Scholar]
- Zouridakis M, Giastas P, Zarkadas E, Chroni-Tzartou D, Bregestovski P, Tzartos SJ. (2014) Crystal structures of free and antagonist-bound states of human alpha9 nicotinic receptor extracellular domain. Nat Struct Mol Biol 21:976–980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.