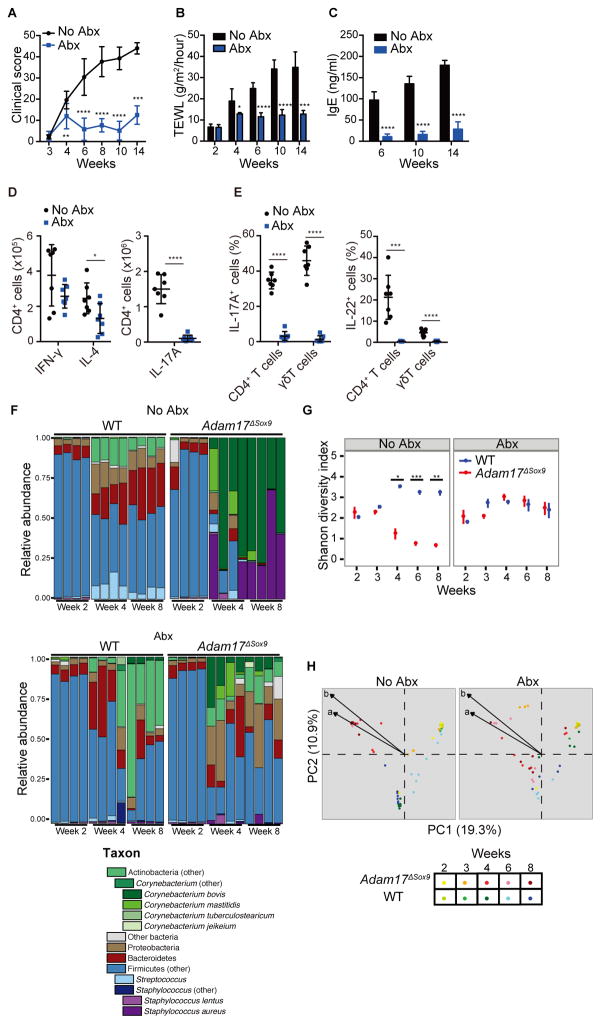
Figure 3. Antibiotics prevent eczematous inflammation and conserves bacterial diversity.
Adam17ΔSox9 mice were either treated or untreated with antibiotics cocktail (Abx) since weaning. See also Figure S3. (A–C) Clinical score, TEWL and serum IgE concentrations (N=7). A C shown as mean ± SD. *P<0.05, **P<0.01, ***P<0.001, ****P <0.0001 as determined by Student’s t test. (D) Flow cytometry analysis of IFN-γ, IL-4 and IL-17A production in CD4+ cells from lymph nodes, and (E) IL-17A and IL-22 production by CD3+ cells in epidermis of WT and Adam17ΔSox9 mice (N=7). (F) Microbiota of Abx-treated and untreated Adam17ΔSox9 and WT mice at indicated time points after birth. Relative abundance plots of order-genera that represented >1% of total 16S rRNA gene sequences with additional speciation of Corynebacterium and Staphylococcus genus of 4 representative mice for each time point. Results are representative of two experiments. (G) Shannon diversity index of Abx-treated and untreated Adam17ΔSox9 and WT mice from week 2 to 8 after birth. Each time point represents mean ± SEM. *P<0.05, **P<0.01, ***P<0.001 determined by Student’s t test and adjusted for multiple comparison by Benjamini and Hochberg correction. (H) Principal coordinates analysis (PCoA) of the microbial community structure (Theta similarity index) of Abx-treated and untreated Adam17ΔSox9 and WT mice. A shorter distance between points indicates higher similarity between the microbiome of samples represented by these points. Biplot arrows indicate the two most significant unique consensus taxonomies contributing to variation along axes 1 and 2 as determined by Spearman correlation. Length of biplot arrows reflects the contribution of that taxa to the top two axes: a, S. aureus (r=−0.73, P<1e-15); b, C. bovis (r=−0.77, P<1e-15). Spearman correlations and P-values refer to axis 1.