Fig 3.
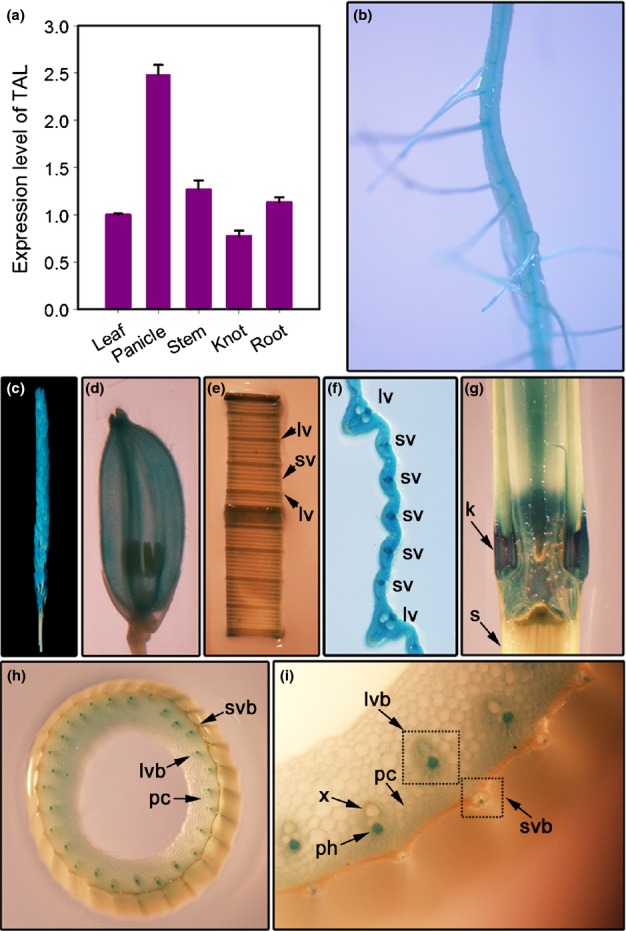
Expression analyses of the rice (Oryza sativa) TAL-type transaldolase (TAL) gene. (a) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of rice TAL expression. Total RNA was isolated from the leaf, panicle, stem, knot, and root of wildtype plants at the heading stage. Amplification of the rice Actin gene was used as a control. Error bars represent + 1SE. (b) Beta-glucuronidase (GUS) activity in root. (c) GUS activity in panicle. (d) GUS activity in glume. (e) GUS activity in leaf at the heading stage. (f) Magnified image of the leaf in (e) showing the strong expression of TAL in developing vascular bundles of leaf. (g) GUS activity in knot and stem. (h) Transverse section of a third internode at the heading stage, showing the expression of TAL promoter-GUS in the vascular bundles of the stem. (i) Magnified image of the stem in (h) showing the strong expression of TAL in the phloem of vascular bundles. lv, large vein; sv, small vein; s, stem; k, knot; lvb, large vascular bundles; svb, small vascular bundles; pc, parenchyma cells; x, xylem; ph, phloem.
