Figure 1.
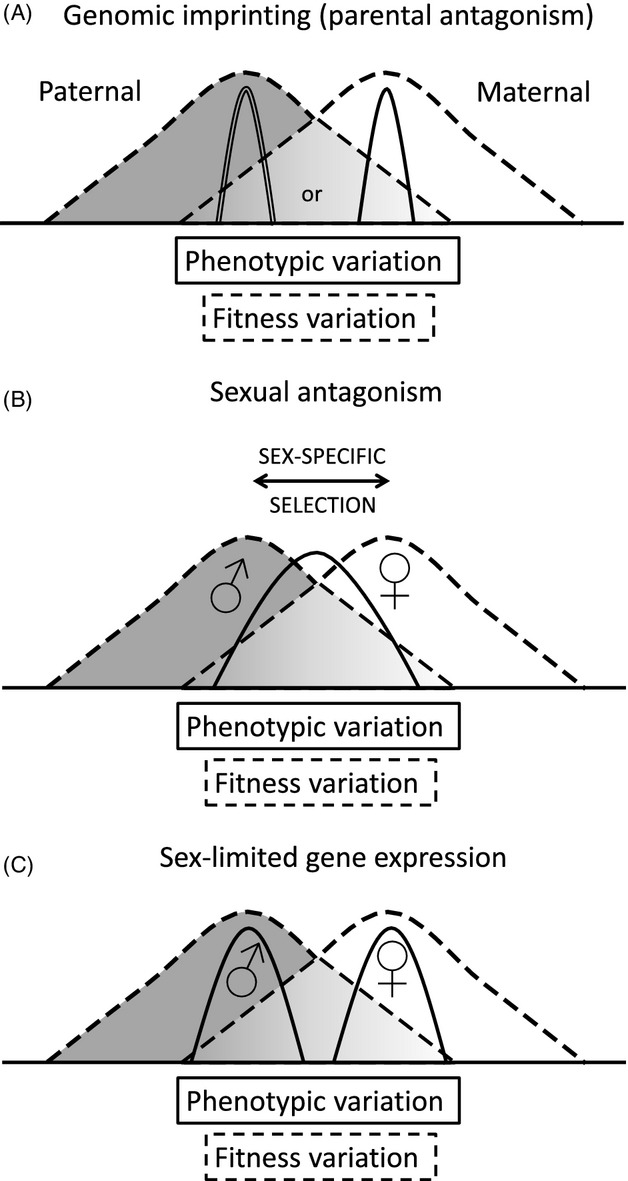
(A) A model of phenotypes mediated by genomic imprinting (parental antagonism). Under imprinting, one parent silences an allele during gametogenesis, thereby gaining a short-term fitness advantage. Subsequent selection on expression of the allele from the other parent leads to optimization of gene expression for that parent, for this locus. Resolution via joint optimization is not feasible in this system, so deleterious effects on health of one or both parties are expected unless multiple paternal and maternal loci exert effects, on the same phenotype, that balance. (B) A model of phenotypes experiencing sexual antagonism. The dashed lines indicate the fitness surfaces for females and males, respectively, and the single solid line indicates the distribution of phenotypic variation for the population. Deviations from sex-specific optima are expected to engender proportional deleterious effects on sex-specific health. (C) Distributions of phenotypic traits resulting from sex-limited gene expression, with sexual antagonism resolved. The dashed lines indicate the fitness surface for females and males, respectively, the single solid line indicates phenotypic variation for the population, with clear sexual dimorphism, and health effects from conflict are not predicted except from new sexually antagonistic mutations.
