Table 2.
Summary of the generalized linear models used for the analysis of the three descriptors of the global pathogen evolutionary trajectories: E, emergence of a crop pathogen (Es = 1 if emergence was successful in simulation s and 0 otherwise); TE, number of years required for the crop pathogen to emerge; 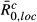 , mean aggressiveness of the pathogen population on the crop at equilibrium
, mean aggressiveness of the pathogen population on the crop at equilibrium
| Descriptor | Distribution | Link function | Linear predictor |
|---|---|---|---|
| Emergence (E) | Binomial | Logit | −1 + LOC + PROP + BETA + CROP + DISP |
| Time before emergence (TE) | Gamma | Log | −1 + LOC + LOC:(PROP + BETA + CROP + DISP + PROP:BETA + PROP:CROP + PROP:DISP + BETA:CROP + BETA:DISP + CROP:DISP) |
Pathogen aggressiveness on the crop (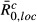 ) ) |
Normal | Natural | −1 + LOC + LOC:(PROP + BETA + CROP + DISP + PROP:BETA + PROP:CROP + PROP:DISP + BETA:CROP + BETA:DISP + CROP:DISP) |
LOC, two-level factor of wild patches spatial aggregation (random or clustered); PROP, scaled wild host proportion; BETA, scaled parameter of the trade-off function (β); CROP, scaled cropping season duration; DISP, scaled pathogen mean dispersal distance; :, interactions.
