Fig. 4.
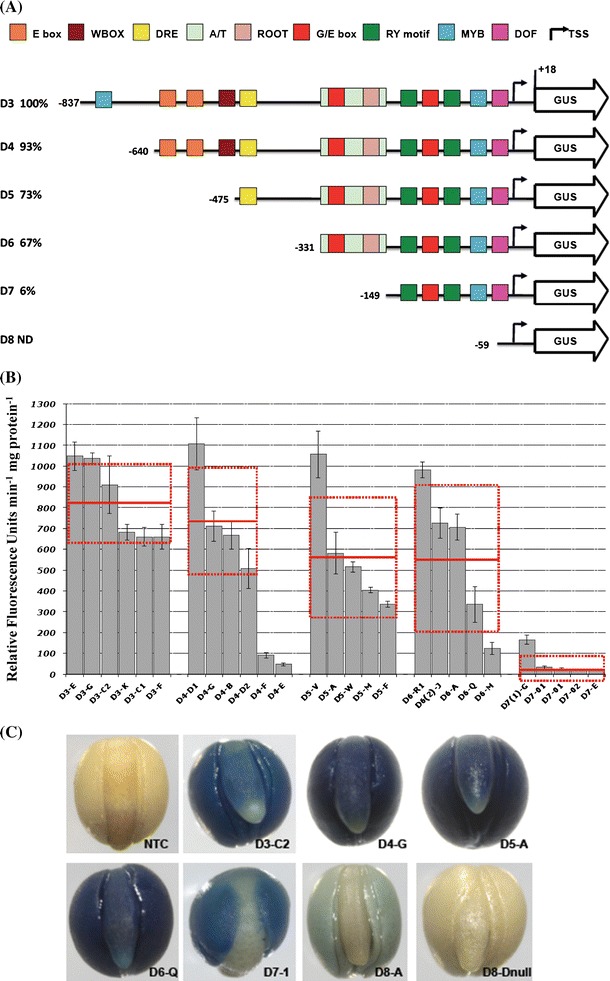
Expression of Bn-FAE1.1 in embryos of B. napus promoter-deletion lines. a Bn-FAE1.1 promoter-deletion constructs. Fragments amplified from the region upstream of the Bn-FAE1.1 coding sequence were used to generate a series of promoter fragments fused to the GUS reporter gene named D3–D8. Positions of deletion end-points relative to the transcription start site are indicated by numbers. Percentage values are relative to activity of D3 deletion. The locations of putative cis-elements are indicated as blocks. ND activity not detected. b Reporter gene activity in Bn-FAE1.1 promoter-deletion lines. GUS activity was quantified in protein extracts isolated from mature seeds. Extracts were prepared from three seeds of four independent plants (indicated by histogram and error bars) and from each of five or six transformation events corresponding to each promoter deletion. Enzyme and protein assays were performed in duplicate for each of the three biological replicates. Red bars represent overall means and standard errors for all plants of each event for each deletion. c Bn-FAE1.1 expression in mature embryos. D3–D8 corresponds to promoter deletion series indicated in a, plants from individual transformation events are identified by letters or numbers after the hyphen. NTC is a non-transformed control plant and null indicates a wild type seed segregating in T1 plants
