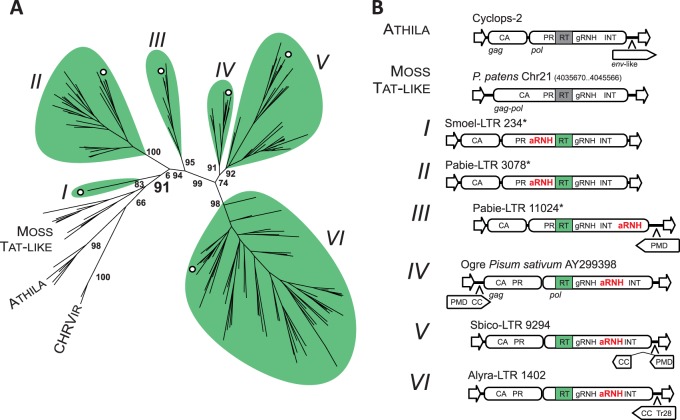
Fig. 2.
Diversity of Tat LTR retrotransposons. (A) Maximum-likelihood tree based on the amino acid sequences of RT from representatives of Tat LTR retrotransposons, Athila elements and chromoviruses. Statistical support was evaluated by the aLRT and is shown at the corresponding nodes of the tree. Six lineages of aRNH-containing Tat LTR retrotransposons are denoted by Latin numerals I–VI and are highlighted in green. Moss Tat-like elements, which are phylogenetically close to aRNH-containing Tat LTR retrotransposons but lack aRNH, are also denoted on the tree, as are Athila elements and chromoviruses (CHRVir). The branches corresponding to representative elements from figure 2B are denoted by white circles. The complete phylogenetic tree with accession numbers and names of the elements is presented in supplementary figure S2, Supplementary Material online. (B) Structural composition of the elements from identified lineages. Schemes of the structures of elements from corresponding lineages are shown. The structure of one of the representative copies was used, with the exception of those denoted by asterisks, for which multiple copies of the element were used to reconstruct the overall structure due to the lack of putatively intact copies. eORFs are denoted by pentagons with the orientation corresponding to the frame (sense/antisense). CA, capsid domain of Gag protein; gRNH, RNH of Ty3/gypsy LTR retrotransposons.