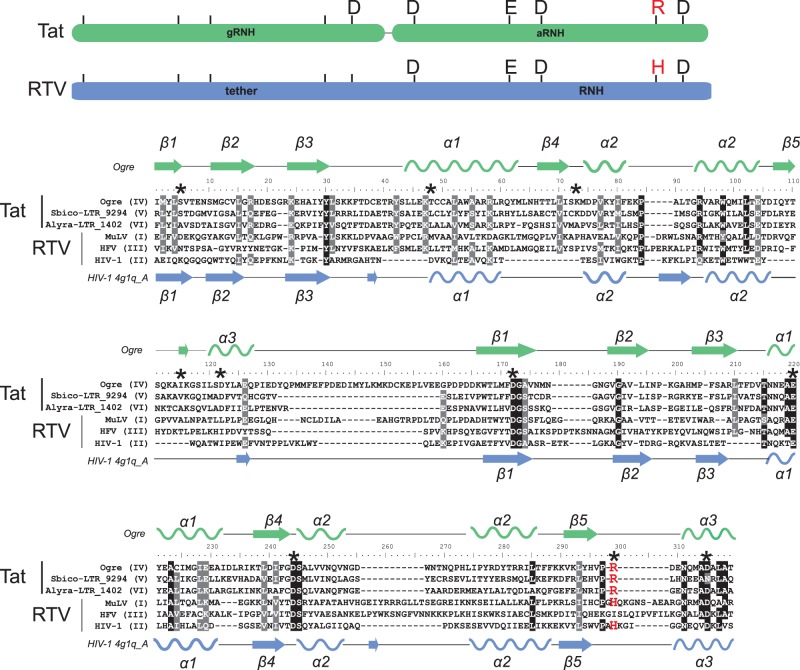
Fig. 4.
Alignment of dual RNHs of diverse Tat LTR retrotransposons and retroviruses (RTV). The names of the sequences used are presented at the left of the alignment and contain information on their lineage for Tat LTR retrotransposons (lineages IV-VI) or their class for retroviruses (classes I-III). Schemes of dual RNHs from Tat LTR retrotransposons (gRNH-aRNH) and retroviruses (tether-RNH) with the indicated active site residues are shown at the top of the figure and are highlighted in green and blue, respectively. The conserved R or H residue of the active site, which is specific for different RNH subtypes, is highlighted in red in the scheme. The secondary structure predicted for gRNH-aRNH of the Ogre element (NCBI accession number AAQ82037) is shown at the top of the alignment. The secondary structure of tether-RNH of HIV-1 retrieved from PDB (accession number 4g1q_A) is shown at the bottom of the alignment. The α-helices are depicted as helices, and the β-sheets are shown as arrows. Positions corresponding to residues of the active site are denoted by black asterisks in the alignment. R or H residues of the active site are highlighted in red.