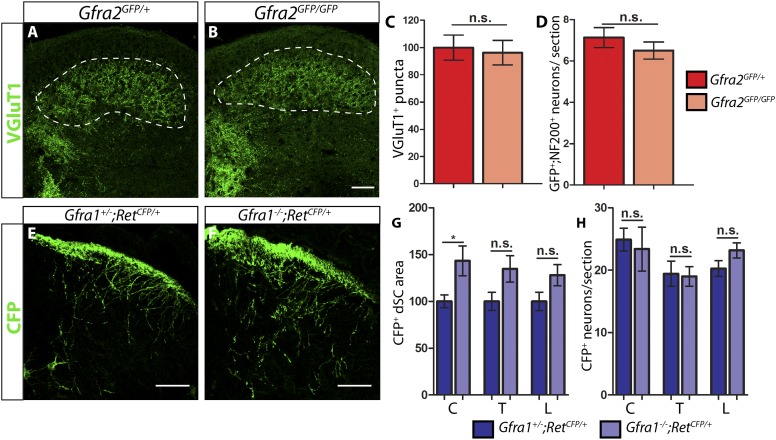
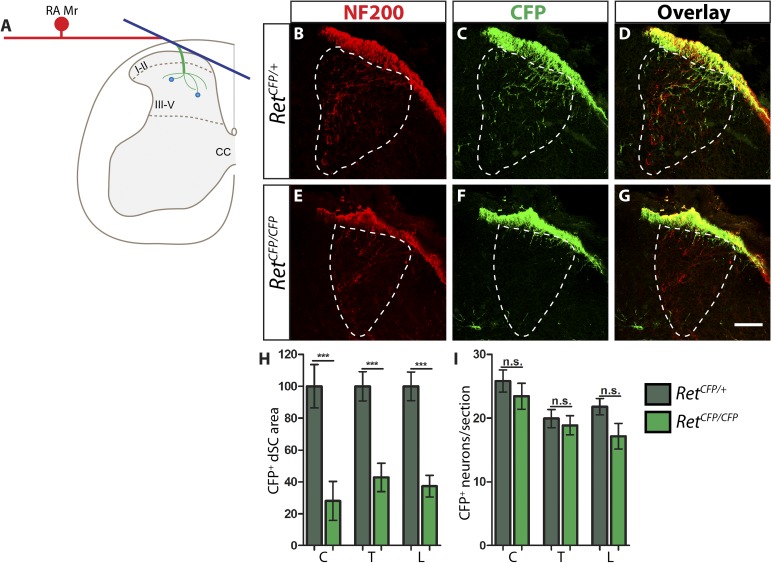
Figure 1. P7 Gfra2 mutant mice show normal dorsal spinal cord (dSC) VGLUT1 staining and Gfra1 null mice display normal rapidly adapting (RA) mechanoreceptor central projections at E13.5.
(A–B) Anti-VGLUT1 immunostaining of P7 SC sections from Gfra2GFP/+ control (A) and Gfra2GFP/GFP null (B) mice. VGLUT1 staining labels presynaptic terminals of mechanosensory neurons, which are found in layers III–V of the dSC (outlined in white). Note that green fluorescent protein (GFP) driven from the Gfra2 locus cannot be visualized directly. Therefore, positive signal indicates presynaptic VGLUT1+ puncta and not GFRa2+ primary afferent axons. (C) Quantification of VGLUT1+ puncta in dSC, which is displayed as a percentage of VGLUT1+ pixels compared to the control pixel count. The similar density of VGLUT1+ puncta between mutant and control tissue suggests that cis RET signaling via GFRa2 is dispensable for the growth of RA mechanosensory central projections at P7. (D) Quantification of GFP+;NF200+ neurons, which indicate RA mechanoreceptors, per DRG section. The non-significant decrease in RA mechanoreceptor number per section in Gfra2 nulls suggests that most RA mechanoreceptors are not dependent on cis RET signaling for survival. (E–F) Anti-GFP immunostaining of RA mechanoreceptor central projections in E13.5 Gfra1+/−;RetCFP/+ control (E) and Gfra1−/−;RetCFP/+ mutant (F) SC sections. The increased CFP signal in Gfra1 null dSC is likely due to the precocious expression of Ret in some dSC neurons of Gfra1 mutants. (G) Quantification of CFP+ pixel number in dSC. The lack of a reduction in CFP+ axons in Gfra1 mutant dSC indicates that trans signaling via GFRa1 is not required for the initial growth of RA mechanosensory third order central projections. (H) Quantification of number of CFP+ neurons per DRG section indicates no loss of RA mechanoreceptors in Gfra1 mutants at E13.5. C: cervical level, T: thoracic level, L: lumbar level. Scale bars = 50 μm. Error bars represent SEM. n.s. = p > 0.05, * = p < 0.05. Source data are provided in Figure 1—source data 1, 2.
DOI: http://dx.doi.org/10.7554/eLife.06828.003