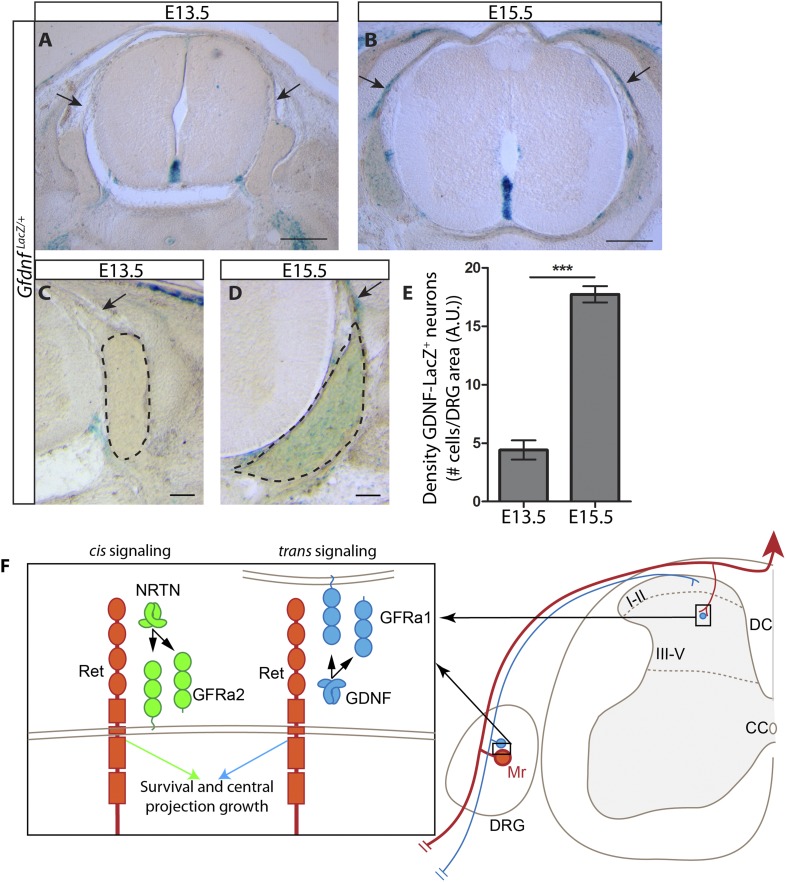
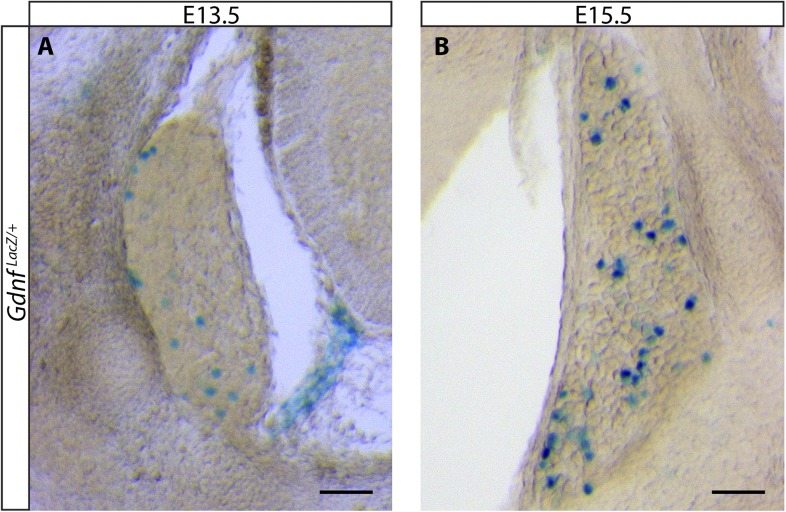
Figure 8. Dynamic expression of GDNF during development.
(A–D) X-Gal staining of E13.5 (A, C) and E15.5 (B, D) GdnfLacZ/+ DRG and SC sections (also see Figure 8—figure supplement 1). Arrows indicate dorsal roots, which express Gdnf at E15.5, but not E13.5. (E) Quantification of LacZ+ cells per DRG section, normalized to DRG area, reveals a significant increase in the number of cells expressing Gdnf from E13.5 to E15.5. E13.5 embryos have 4.41 ± 0.82 LacZ+ cells/unit area of DRG, E15.5 embryos have 17.73 ± 0.70 LacZ+ cells/unit area of DRG (p < 0.001). Error bars represent SEM. Scale bars = 200 μm (A–B), 100 μm (C–D). *** = p < 0.001 (F) Model of cis and trans signaling at cell bodies and central branches of RA mechanoreceptors. GFRa2 is co-expressed with RET in RA mechanoreceptors and can activate RET in cis. GFRa2 can also be shed from the membrane and may activate RET in its soluble form. GFRa1 is expressed in neighboring DRG neurons, dorsal root entry zone cells, and dSC cells. GFRa1 present at the membrane of these cells may directly contact the cell bodies or processes of RA mechanoreceptors to activate RET in trans. In addition, soluble GFRa1 released from these cells may also activate RET in RA mechanoreceptor in trans.