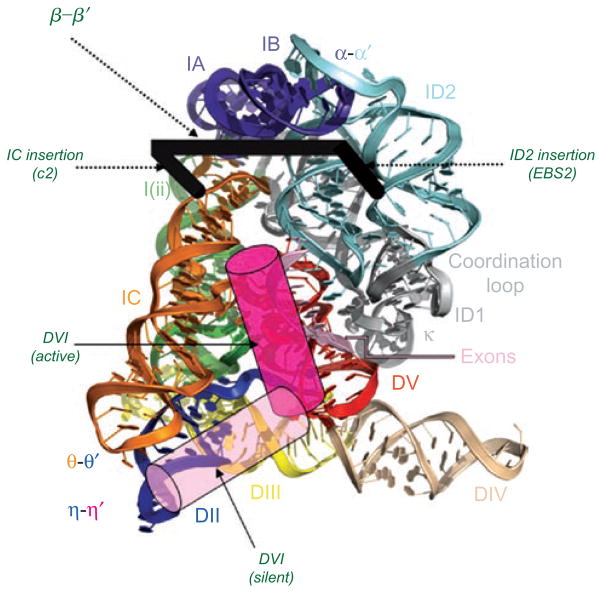
Figure 13.
The hypothetical position of DVI, EBS2 and β–β′. Superimposed on a model of the group II intron crystal structure (rendered from pdb file 3IGI), the proposed position of various derived domains is shown. DVI is likely to have two functional conformations. The “active” conformation that participates in branching is likely to be in the position indicated by the dark pink cylinder. The “silent” conformation is stabilized by the η–η′ interaction between DVI and DII is likely to be located in the position indicated by the light pink cylinder. DVI probably transits between these two sites in order to toggle between branching and other aspects of intron function (see text). The likely sites of EBS2, β–β′ and the d2a and c2 stems are shown by the thick black bracket. The ID2 insertion (which evolved into EBS2 and the d2a stem) occurred approximately between nucleotides 114 and 115 (Oceanobacillus iheyensis numbering), placing EBS2 on the right hand side of the bracket. The IC insertion occurred approximately between nucleotides 158 and 159, placing helix c2 on the left hand side of the bracket. The β–β′ interaction between loops of c2 and d2a is shown as the connecting bar of the bracket, spanning structural domains of the core.