Figure 4.
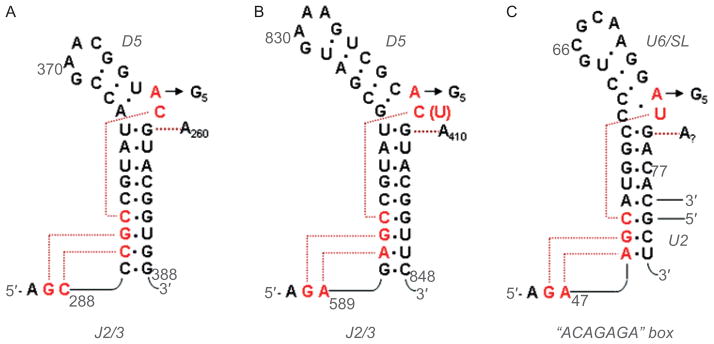
Secondary structures of DV and the U6 snRNA, together with anticipated contacts within the active-site. (A) The DV and J2/3 sequences from group IIC intron Oceanobacillus iheyensis, with active-site tertiary interactions determined from the crystal structure (Keating et al., 2010). The J2/3 nucleotides form a major groove triplex with the “catalytic CGC triad” of conserved nucleotides at the base of DV, and with C259 from the bulge. (B) The DV and J2/3 sequences from group IIB intron ai5γ. Anticipated long-range interactions are shown, by analogy to the O.i. crystal structure. These can be readily modeled into the active-site of the solved structure (Keating et al., 2010). (C) The U6 snRNA from the human spliceosome. The ACAGAGA box is hypothesized to function like J2/3, forming a triple helix that supports the metal-binding, actives-site bulge at positions 73 and 74. The U6 bulge region is hypothesized to form tertiary interactions similar to those observed within the IIC intron (Keating et al., 2010). Reprinted with permission from RNA (Keating et al., 2010).
