Figure 6.
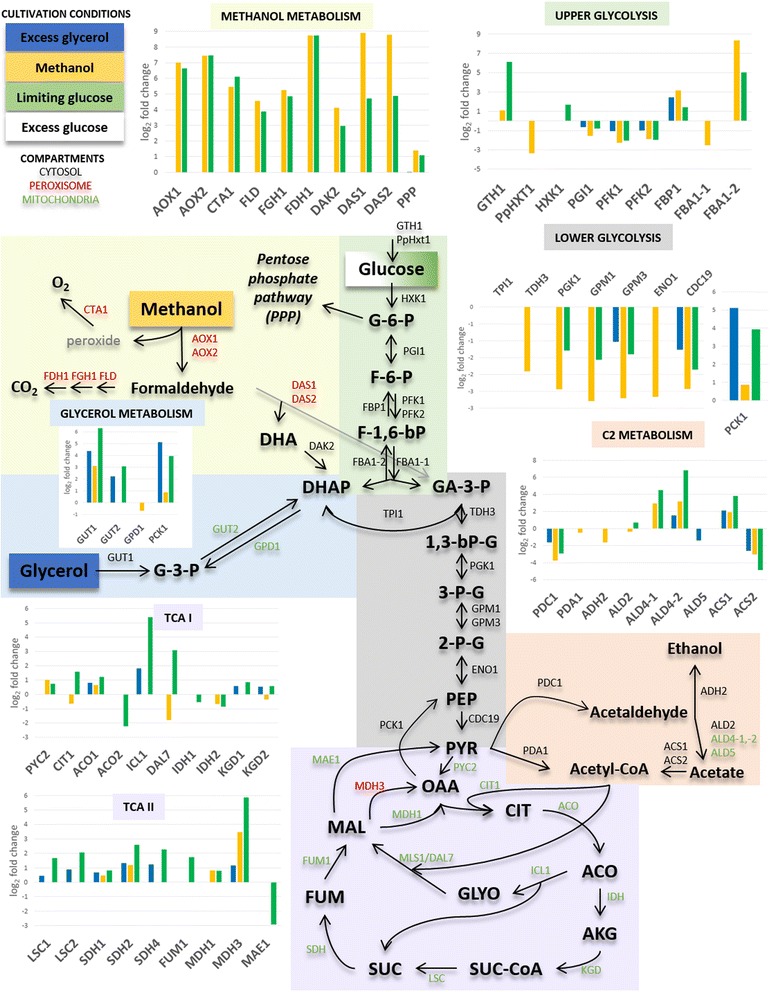
Central carbon metabolism pathways in Pichia pastoris. Transcriptional log2 fold changes of genes significantly regulated in excess glycerol, methanol and limiting glucose compared to excess glucose are presented in bar charts (cutoff ±50% fold change and adjusted p-values < 0.05; [23]). According to cellular localization, peroxisomal, cytosolic and mitochondrial enzymes are colored in red, black and green, respectively. Metabolites: G-6-P: glucose 6-phosphate; F-1,6-P: fructose 1,6-phosphate; DHA(P): dihydroxy acetone (phosphate); G-3-P: glycerol 3-phosphate; GA-3-P: glyceraldehyde 3-phopshate; 1,3-bPG: 1,3-bisphosphoglycerate; 3-PG: 3-phosphoglycerate; 2-PG: 2-phosphoglycerate; PEP: phosphoenolpyruvate; PYR: pyruvate; OAA: oxaloacetate; CIT: citrate; ICIT: isocitrate; AKG: alpha-keto glutarate; SUC: succinate; SUC-CoA: succinyl-Coenzyme A; FUM: fumerate; MAL: malate; GLYO: glyoxylate; Enzymes: AOX1/2: alcohol oxidase; CTA1: catalase A; FLD: bifunctional alcohol dehydrogenase and formaldehyde dehydrogenase; FGH1: S-formylglutathione hydrolase; FDH1: formate dehydrogenase; DAK2: dihydroxyacetone kinase; DAS1/2: dihydroxyacetone synthase; GUT1: glycerol kinase; GUT2: glycerol-3-phosphate dehydrogenase; GPD1: glycerol-3-phosphate dehydrogenase; PCK1: phosphoenolpyruvate carboxykinase; GTH1: high-affinity glucose transporter; HXT1: low-affinity glucose transporter; HXK1: hexokinase; PGI1: phosphoglucose isomerase; PFK1/2: phosphofructokinase; FBP1: fructose-1,6-bisphosphatase; FBA1-1/1-2: fructose 1,6-bisphosphate aldolase; TPI1: triose phosphate isomerase; TDH3: glyceraldehyde-3-phosphate dehydrogenase; PGK1: 3-phosphoglycerate kinase; GPM1/3: phosphoglycerate mutase; ENO1: enolase I, phosphopyruvate hydratase; CDC19: pyruvate kinase; PDC1 pyruvate decarboxylase; PDA1: E1 alpha subunit of the pyruvate dehydrogenase (PDH) complex; ALD2: cytoplasmic aldehyde dehydrogenase; ALD4-1/4-2/5: mitochondrial aldehyde dehydrogenase; ACS1/2: acetyl-coA synthetase; PYC2: pyruvate carboxylase; CIT1: citrate synthase; ACO1/2: aconitase; ICL1: isocitrate lyase; DAL7: malate synthase; IDH1/2: isocitrate dehydrogenase; KGD1: alpha-ketoglutarate dehydrogenase complex; KGD2: dihydrolipoyl transsuccinylase; LSC1: succinyl-CoA ligase; SDH1/2/4: succinate dehydrogenase; FUM1: fumarase; MDH1: mitochondrial malate dehydrogenase; MDH3: malate dehydrogenase; MAE1: mitochondrial malic enzyme.
