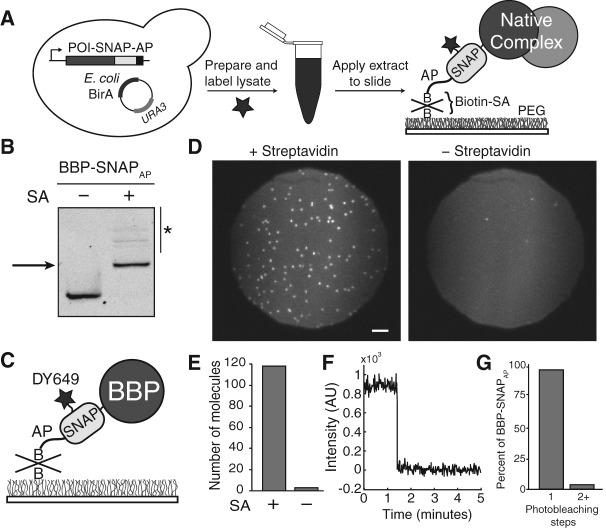
FIGURE 1.
Overview and characterization of SNAP-SiMPull. (A) Schematized workflow of a SNAP-SiMPull experiment. A yeast strain is constructed containing a SNAPAP-derivitized protein. A whole-cell lysate is then prepared, labeled with a benzylguanine fluorophore, and passed through a gel filtration column before application to a glass slide. This results in immobilization of complexes containing the SNAP-tagged protein. (SA) Streptavidin, (B) biotin. (B) Streptavidin EMSA to determine the extent of biotinylation of the SNAPAP tag. The BBP protein was derivitized with a fluorophore prior to SDS-PAGE and fluorescence imaging. Addition of streptavidin causes a decrease in gel mobility indicative of biotinylation of the SNAPAP tag. Asterisk indicates the presence of higher-order streptavidin oligomers binding to fluorescent BBP-SNAPAP. (C) Cartoon representation of DY649-labeled BBP-SNAPAP pull-down on the surface of a passivated glass slide. (D) Representative microscopic fields of view of a SNAP-SiMPull assay with DY649 fluorophore-labeled BBP-SNAPAP. Pull-down is strongly dependent on the presence of streptavidin. Scale bar is 5 μm. (E) Quantification of the number of DY649 BBP-SNAPAP spots observed in the presence and absence of streptavidin coating from representative experiments. (F) Representative single-molecule trace highlighting single step photobleaching for immobilized BBP-SNAPAP molecules. (G) Quantification of the number of observed photobleaching steps for spots of BBP-SNAPAP fluorescence. The vast majority of spots photobleach in a single step consistent with the presence of single molecules of DY649 BBP-SNAPAP on the surface.