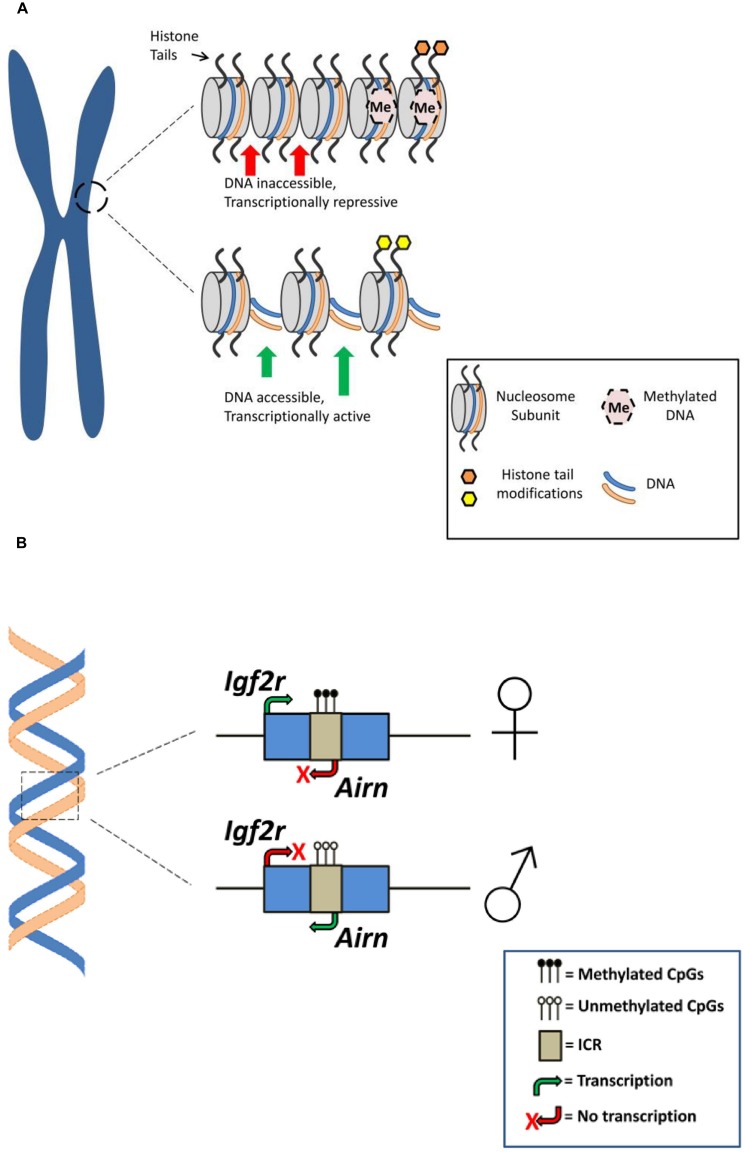
FIGURE 1.
Epigenetic mechanisms associated with genomic imprinting. (A) Histone modifications and DNA methylation for different chromatin configurations. Top: Repressive chromatin state associated with histone modification (e.g., histone methylation; orange shading) and dense DNA methylation resulting in gene silencing or attenuated gene expression. Bottom: Active/permissive chromatin state associated with histone modification (e.g., histone acetylation; yellow shading) and reduced DNA methylation rendering DNA accessible for transcription resulting in gene expression (for a comprehensive overview of histone modifications see Bannister and Kouzarides, 2011). (B) Genomic arrangement at an imprinted gene. A simplified schematic of the murine Igf2r locus demonstrating parent-of-origin specific DNA methylation is presented. The imprinting control region (ICR) on the maternal Igf2r allele is methylated, preventing expression of an antisense ncRNA (Airn) and resulting in expression of the maternal Igf2r allele. Alternatively, expression of Airn from the unmethylated paternal allele attenuates paternal Igf2r expression (for a more comprehensive overview of DNA methylation at the Igf2r locus and genomic imprinting see Autuoro et al., 2014).