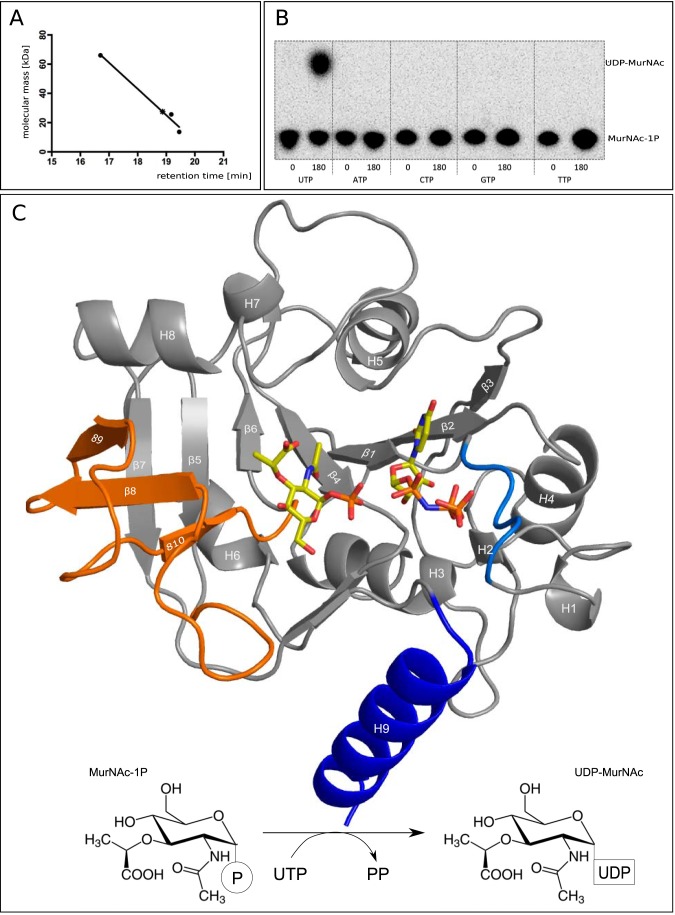
FIGURE 1.
A, HPLC analysis confirms a monomeric state of MurU. Retention times obtained from size exclusion chromatography for the standard proteins albumin (66 kDa), chymotrypsinogen (25.6 kDa), and ribonuclease (13.7 kDa) (filled dots) were plotted against their molecular mass. The retention time for MurU and its corresponding apparent molecular mass (18.87 min and 27.5 kDa) are indicated by an asterisk. B, NTP substrate specificity assay using radiolabeled MurNAc α-1-[32P]phosphate. C, overall structure of a ternary complex of MurU bound to its substrate MurNAc-α1-P and the substrate analog UppNHp. Ligand positions are based on two different crystals that were each soaked with MurNAc-α1-P and UppNHp or UpNHpp an MgCl2, respectively (see Table 1). Substrates are depicted as sticks and colored according to the atom type (oxygens in red, nitrogens in blue, carbons in yellow, and phosphorus in orange). The protein is shown in schematic representation with the colors highlighting the subdomains referred to in the text (blue, C-terminal helix; orange, sugar-binding domain; marine blue, nucleotidyltransferase signature motif). Secondary structure elements are numbered from the N to C terminus. Bottom, schematic representation of the reaction catalyzed by MurU.