FIGURE 2.
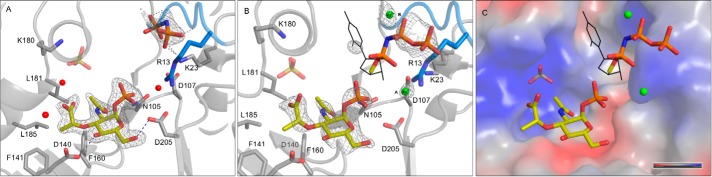
A and B, crystal structures of MurU soaked with MurNAc-α1-P + UppNHp (A; complex 1) and MurNAc-α1-P + UppNHp and MgCl2 (B; complex 2) with omit difference maps (mFo − DFc) for the ligands contoured at 2.5 σ. Parts of the substrates for which omit difference electron density could be obtained are depicted as sticks, Mg2+ ions are depicted as green spheres, and those parts of UpNHpp for which no or only 2mFo − DFc density could be obtained are represented as black lines. The SNT signature motif is highlighted in marine blue. Side chains that are involved in ligand coordination are represented as sticks. Ligands and side chains are colored by atom type (oxygens in red, nitrogens in blue, phosphorus in orange, sulfur in gold, magnesium in green, and carbons in the colors used for the domains in Fig. 1. C, surface representation of the substrate-binding site colored according to the surface potential (gradient from blue (positive) to red (negative)). Note that the parts of UpNHpp for which no reliable density could be obtained were modeled in the most likely possible conformation by relying on geometrical restraints and the clear electron density for the pyrophosphate part as an anchor point.
