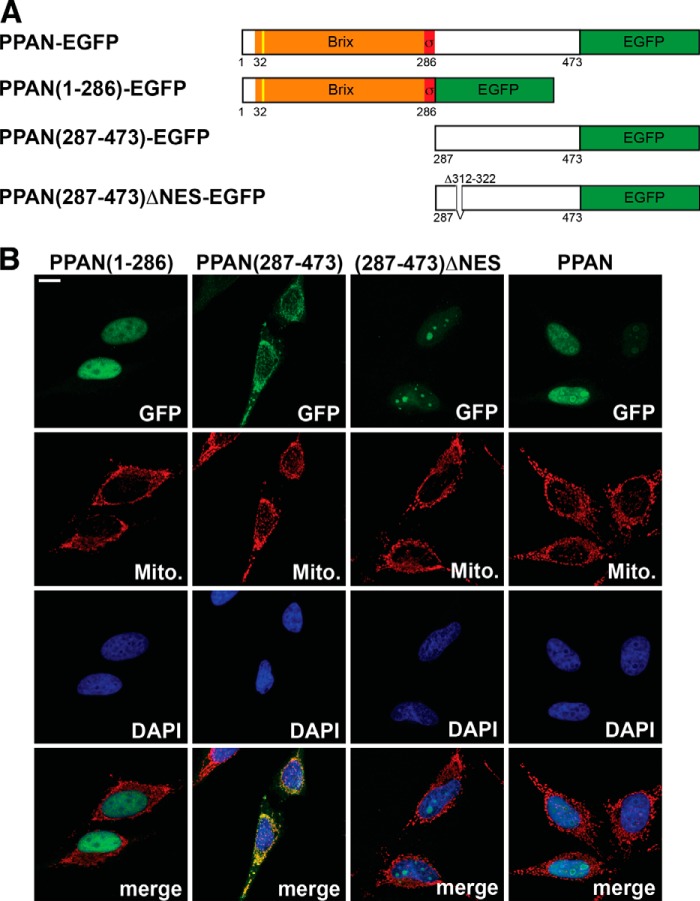
FIGURE 2.
PPAN has a functional NES motif and localizes to mitochondria via its C terminus. A, schematic representation of the PPAN constructs fused to EGFP as depicted. The Brix domain is shown in orange; the σ70-like motif is shown in red, and the EGFP tag is in green. The yellow line indicates the predicted cleavable mitochondrial presequence (cleavage site IRQLS). Numbers show amino acid positions. NES, nuclear export sequence. Δ312–322 denotes the internally deleted NES. B, subcellular localization of PPAN constructs when overexpressed. PPAN constructs were transiently transfected in HeLa cells as shown above panels and stained for GFP, mitochondria (Mito.), and nuclei (DAPI) as depicted within the panels. Of note, the C terminus of PPAN(287–473) localizes to mitochondria, whereas the corresponding ΔNES mutant accumulates in the nucleoli. Scale bar, 10 μm. Fluorescence images are representative of four independent experiments.