FIGURE 1.
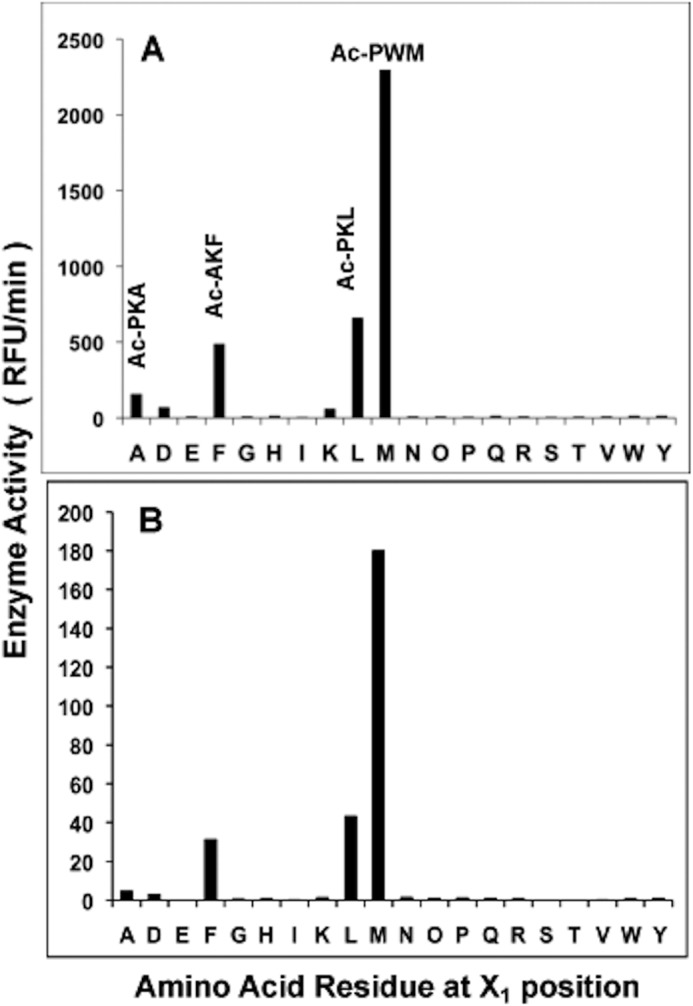
Mtb ClpP1P2 cleaves peptide-amc substrates mainly after Leu, Phe, Ala, and especially Met. A, to determine cleavage preferences for the X1 position, ClpP1P2 activity was measured continuously using Ac-X3X2X1-amc fluorogenic peptides library (at 10 μm) in buffer A containing 30 nm ClpP1P2. The ClpP1P2 activity against the best substrates with a given amino acid in X1 position is reflected in the graph. B, average rate of hydrolysis of substrates with a given amino acid in the X1 position. The activity was measured as in Fig. A. RFU, relative fluorescence units.
