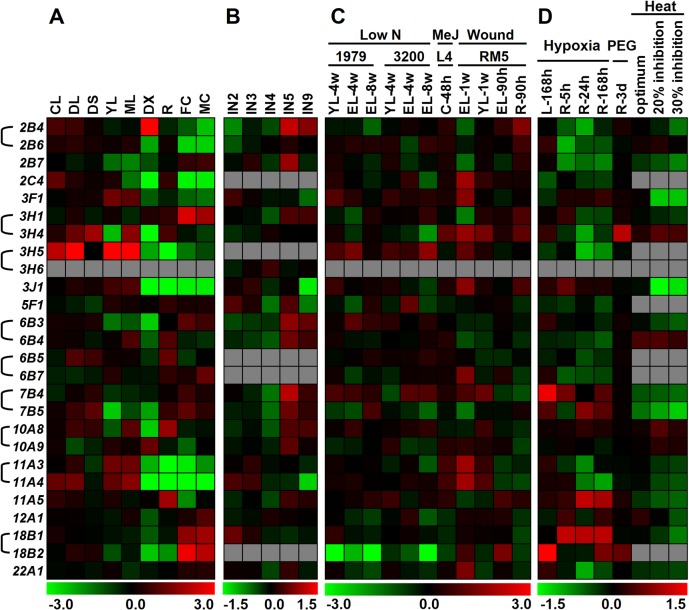
Fig 4. Expression profiles of Populus ALDH genes across different tissues and various abiotic stresses.
(A) Heatmap showing expression profiles of PtALDH genes across various tissues analyzed. The Affymetrix microarray data were obtained from NCBI Gene Expression Omnibus (GEO) database under the series accession number GSE13990. CL, continuous light-grown seedling; DL, etiolated dark-grown seedling transferred to light for 3 h; DS, dark-grown seedlings; YL, young leaf; ML, mature leaf; DX, differentiating xylem; R, root; FC, female catkins; MC, male catkins. (B) Heatmap showing expression profiles of PtALDH genes at different stem development stages. The NimbleGen microarray data were obtained from NCBI GEO database under the series accession number GSE17230. IN2-IN9, stem internodes 2 to stem internodes 9. (C) Heatmap showing expression profiles of PtALDH genes across various stresses and genotypes analyzed. Microarray data under the series accession number GSE16786 was obtained from NCBI GEO database. Genotypes analyzed included: P. fremontii × angustifolia clones 1979, 3200, and RM5, P. tremuloides clones 271 and L4, and P. deltoids clones Soligo and Carpaccio. Tissues analyzed included: YL, young leaves; EL, expanding leaves; R, root tips; C, suspension cell cultures. Stress treatments included: low N, nitrogen limitation; MeJ, Methyl Jasmonate elicitation; Wound, sampled either one week or 90 hours after wounding. (D) Heatmap showing expression profiles of PtALDH genes under hypoxia, drought and heat stresses. Microarray data under the series accession number GSE13109 (hypoxia), GSE17225 (drought) and GSE26199 (heat) was obtained from NCBI GEO database. Stress treatments included: Hypoxia, the root system of grey poplar (Populus × canescens) were flooded for up to 168 h; Drought, a moderate water deficit was applied by adding PEG to the nutrient solution (200g/l) on hybrid poplar (P. deltoides × P. nigra) grown in hydroponics; Heat, fully expanded leaf samples of P. trichocarpa were harvested at 4 physiological states as determined from prior gas exchange measurements (growth temperature, 22°C—baseline, 31.75°C—photosynthetic optimum, 38.4°C—20% inhibition of optimum and 40.5°C—30% inhibition of optimum). Color scale represents log2 expression values, green represents low level and red indicates high level of transcript abundances.