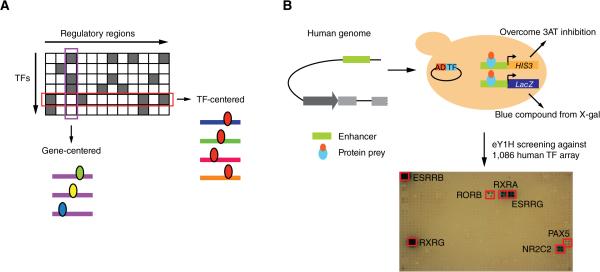
Figure 1. Gene-Centered Yeast One-Hybrid Assays.
(A) Gene-centered versus TF-centered approaches for mapping protein-DNA interactions. Rectangles – regulatory regions; ellipses – TFs.
(B) Cartoon of eY1H assays. A DNA sequence of interest is cloned upstream of two reporter genes (HIS3 and LacZ) and integrated into the yeast genome (i.e., each DNA bait is tested in duplicate by activation of each reporter in the same yeast nucleus). The resulting yeast DNA bait strain is mated to a collection yeast strains harboring TFs fused to the Gal4 activation domain (AD). Positive interactions are determined by the ability of the diploid yeast to grow in the absence of histidine and overcome the addition of 3AT a competitive inhibitor of the HIS3 enzyme, and turn blue in the presence of X-gal. Each TF is tested in quadruplicate. Red boxes show positive interactions.