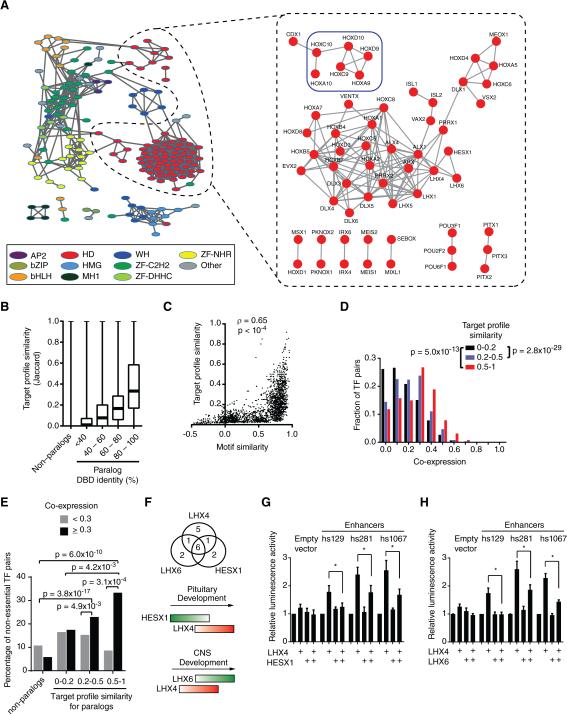
Figure 3. TF Redundancy and Opposing Functions.
(A) TF association network. Each node represents a TF and edges connect TFs with a target profile similarity ≥0.2 (left, all TF families) or ≥0.45 (right, homeodomains). TFs with degree ≥ 3 in the eY1H network are shown. Node color indicates TF families. Colored squares highlight sets of TFs discussed in main text. AP2 – activating protein 2; bZIP – Basic Leucine Zipper Domain; bHLH – basic helix-loop-helix; HD – homeodomain; HMG – High-Mobility Group; MH1 – Mad homology 1; WH – Winged Helix; ZF-C2H2 – Zinc Finger C2H2; ZF-DHHC – Zinc Finger DHHC; ZF-NHR – Nuclear Hormone Receptor.
(B) Target profile similarity between TFs according to DNA binding domain identity. For each pair of TF paralogs with different DNA binding domain amino acid identity their target profile similarity was determined. Each box spans from the first to the third quartile, the horizontal lines inside the boxes indicate median value and the whiskers indicate minimum and maximum values. All pairwise comparisons between groups are significant (p < 0.01) by Dunn's multiple comparison test.
(C) Correlation between motif similarity and target profile similarity. For each TF pair target profile similarity was plotted against their DNA motif similarity determined as the Pearson correlation coefficient of the Z-scores obtained for all possible 8-mers in protein binding microarrays.
(D) Histogram of spatiotemporal co-expression for TF pairs according to their target profile similarity. Statistical significance determined by Mann-Whitney's U tests.
(E) Redundancy between TFs. Each pair of TF paralogs was binned according to their target profile similarity and according their spatiotemporal co-expression. The percentage of TF-pairs for which both TF knockouts are viable was determined. Statistical significance was determined using the proportion comparison test.
(F) Top: overlap between enhancers bound by LHX4, LHX6 and HESX1. Bottom: cartoon of developmental expression. Red – transcriptional activator; green – transcriptional repressor.
(G) HESX1 represses LHX4-induced enhancer activity. HEK293T cells were co-transfected with enhancer constructs cloned upstream of a Firefly luciferase reporter vector, and the indicated TF expression vectors. After 48 hrs, cells were harvested and luciferase assays were performed. Relative luminescence activity is plotted as fold change compared to cells co-transfected with control vector expressing GFP. Experiments were performed three times in 3-6 replicates. Average relative luminescence activity ± SEM is plotted. *p<0.05 by Student's t-test.
(H) LHX6 represses LHX4-induced enhancer activity. Experiments were performed three times in 3-6 replicates. Average relative luminescence activity ± SEM is plotted. *p<0.05 by Student's t-test.
See also Figures S2 and S3.