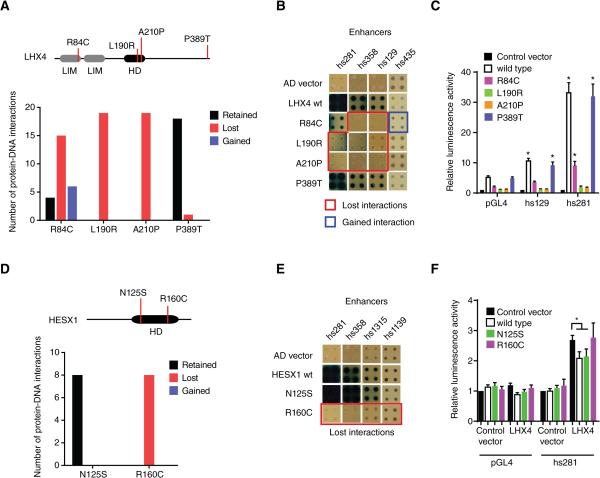
Figure 5. Disease-Associated Coding Mutations in TFs.
(A) Four missense mutations in LHX4 were tested for loss or gain of protein-DNA interactions in eY1H assays against 152 enhancers. The top panel depicts a cartoon of LHX4, including the location of the mutations and the homeodomain (HD) and LIM domains. The bottom panel shows the number of interactions retained (black bar), lost (red bar) or gained (blue blue) for each mutant compared to wild type interactions.
(B) Examples of interactions lost and gained for LHX4 missense mutations. Each TF-enhancer combination was tested in quadruplicate three times. One random quadruplicate test is shown corresponding to four enhancers. Red squares – interaction lost with TF mutant; blue square – retained interaction with TF mutant; AD vector – empty prey vector.
(C) Transcriptional activation mediated by wild type and mutant LHX4 alleles. HEK293T cells were co-transfected with enhancer constructs cloned upstream of a Firefly luciferase reporter vector, and the indicated TF expression vectors. Relative luminescence activity is plotted as fold change compared to cells co-transfected with empty expression vector. Experiments were performed four times with three replicates each. Average relative luminescence activity ± SEM is plotted. *p<0.05 vs empty expression vector by Student's t-test.
(D) Two missense mutations in HESX1 were tested for changes in protein-DNA interactions as in (A).
(E) Examples of interactions lost for HESX1 missense mutations.
(F) Repression of LHX4-induced enhancer activity by wild type and mutant HESX1 alleles. HEK293T cells were co-transfected with enhancer constructs cloned upstream of a Firefly luciferase reporter vector, and the indicated TF expression vectors. Relative luminescence activity is plotted as fold change compared to cells co-transfected with control vector expressing GFP. Experiments were performed six times with three replicates each. Average relative luminescence activity ± SEM is plotted. *p<0.05 by Student's t-test.
See also Table S4.