Figure 1.
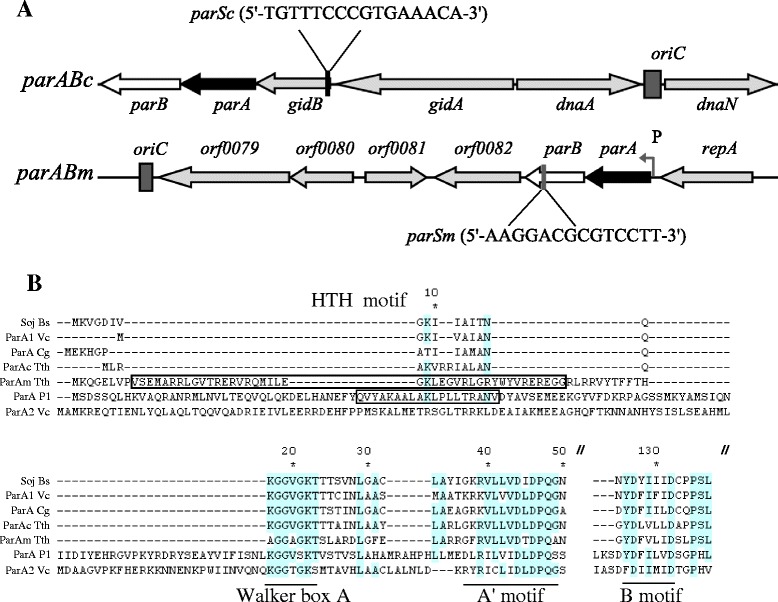
Organization of the chromosomal and megaplasmid par loci, and features of the corresponding ParA proteins. (A) Schematic organization of the parABc and parABm regions. Genes encoding proteins with unknown functions are labeled with their ORF numbers. The predicted replication origins in the chromosome and megaplasmid are indicated with dark-gray bars. The positions and sequences of the chromosomal and megaplasmid parS sites are shown. (B) Part of a multiple sequence alignment of T. thermophilus ParAc, ParAm and ParA proteins from representative bacterial species. Abbreviations: Bs, B. subtilis; Vc, V. cholerae; Cg, Corynebacterium glutamicum; Tth, T. thermophilus; P1, E. coli P1 phage. Completely conserved amino acids are shown with blue color. Black frames indicate the predicted helix-turn-helix (HTH) motifs identified in P1 ParA, and Tth ParAm.
