Figure 5. Asymmetric Nucleosome Unwrapping Controlled by DNA Local Flexibility.
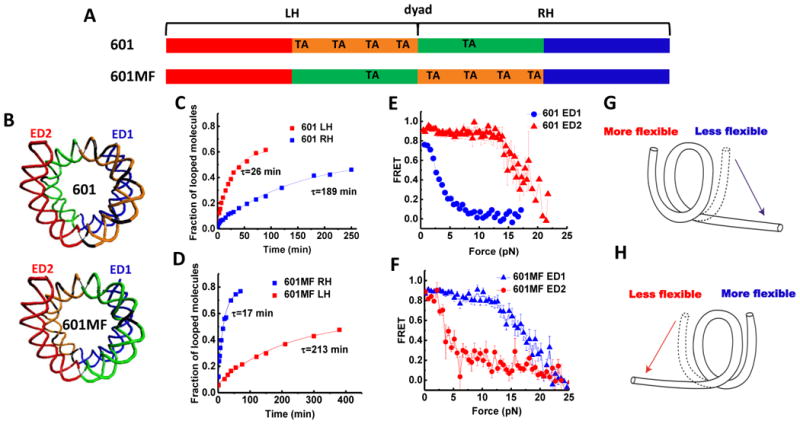
(A) Variations of the 601 the sequence where the inner quarters are colored in orange and green and the outer quarters are colored in red and blue. TA steps are indicated. (B) Nucleosomal DNA structures are shown in the same color scheme with corresponding scheme of the sequence. (C and D) Single exponential fits to the looped DNA fraction versus time yield the average looping time τ measured using single DNA cyclization assay for the 73 bp left or right halves (LH and RH, respectively). (E and F) Averaged stretching time traces of FRET efficiency versus force for nucleosomes in ED1 and ED2 labeling schemes. Error bars denote SEM of 25 traces for 601 ED1, 15 traces for 601 ED2, 29 traces for 601MF ED1, 19 traces for 601MF ED2. (G and H) Illustrations of the relationship between the direction of nucleosome unwrapping and the DNA flexibility of the two halves of the nucleosomal DNA sequence. The nucleosome unwraps from the stiffer side (single-headed arrows) if the DNA flexibility differs significantly between the two sides. See also Figure S5.
