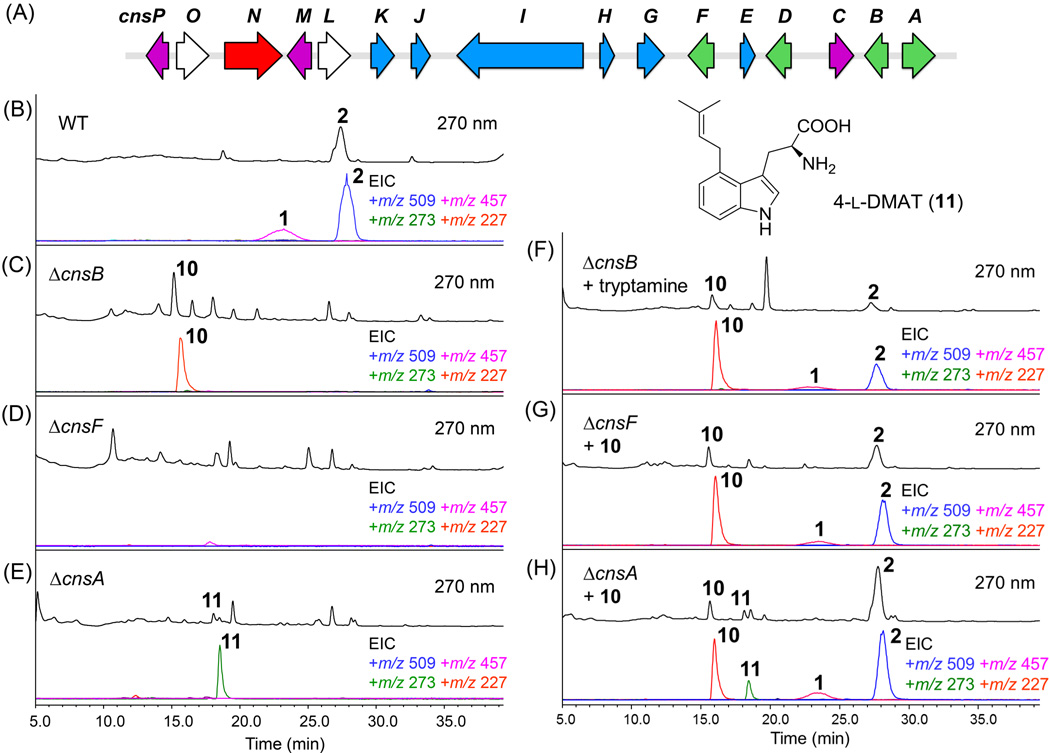
Figure 2.
Mapping the early stages of the communesin biosynthetic pathway. (A) The putative cns gene cluster from P. expansum NRRL 976. Genes are color-coded according to functions of encoded enzymes: precursor synthesis (green), post-core tailoring modifications (blue), other oxygenases (purple), fungal transcription factor (red) and those with unknown/unassigned function (white) (see Table S1). (B) Confirmation of production of 1 and 2 from the wild type strain. The UV (λ=270 nm) trace is shown above and the selected ion monitoring MS trace is shown below with the masses indicated in diffrent colors; (C)–(E) LC-MS analysis of metabolites extracted from ΔcnsA, ΔcnsB and ΔcnsF. (F)–(H) LC–MS analysis of metabolites produced from mutant strains shown in (C)–(E) after chemical complementation with proposed biosynthetic precursors. Tryptamine (F) or 10 (G and H) was supplemented at ~40 µg/mL.