Figure 4.
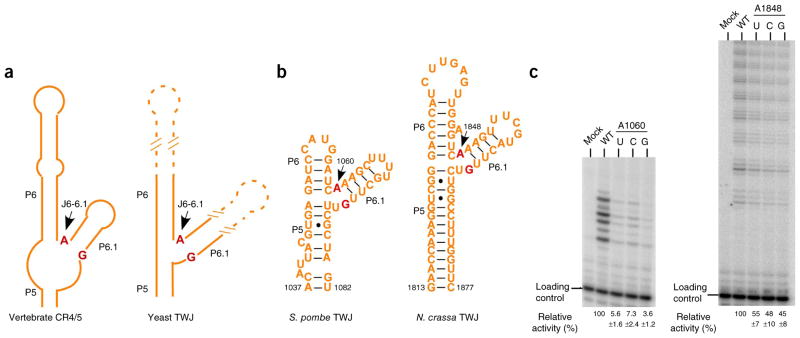
Phylogenetic and functional analysis of the invariant junction J6-6.1 nucleotide A. (a) Schematic diagram of the secondary structures of vertebrate CR4/5 and yeast TWJ. The structure-based sequence alignment is shown in Supplementary Figure 2b. The invariant nucleotide A at the junction J6-6.1 and its pairwise nucleotide G are colored in red. (b) Secondary structures of S. pombe and N. crassa TWJ domains. (c) Telomerase primer-extension assays of wild-type and mutant S. pombe and N. crassa telomerases (additional data in Supplementary Fig. 4c). Activities of mutant telomerases (A1060U, A1060C and A1060G substitutions for S. pombe and A1848U, A1848C and A1848G substitutions for N. crassa) are normalized to the wild type (WT) and shown below the gel as relative activities ± s.d. from three independent assays.