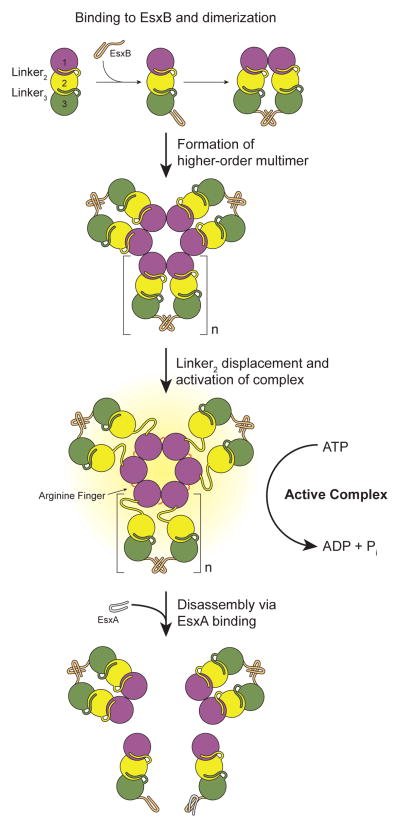
Figure 7. Model of substrate-mediated activation of EccC.
In the absence of substrates EccC is monomeric. Interaction with EsxB leads to dimerization of the ATPase and then higher order multimerization, but cannot activate the enzyme. In this study we used the R543A mutation to disrupt the interaction between ATPase1 and ATPase2 although in vivo this role may be played by other proteins that bind to ATPase1 analogously to the binding of EsxB to ATPase3, or other signals. Once ATPase1 is displaced, EccC is activated further by multimerization mediated by a conserved R-finger. EsxA can disrupt the EsxB:EsxB interaction and disassemble the multimer. We have no evidence for the structure of the EccC:EsxB dimer or stiochiomtry and structure of the multimeric form. Thus, both these aspects of the model are speculative, though based on prior structures of related substrate proteins (i.e. 3GVM) and the FtsK-like ATPases (i.e. 2IUU). We have indicated this ambiguity with the variable “n” for the number of subunits in the multimer.