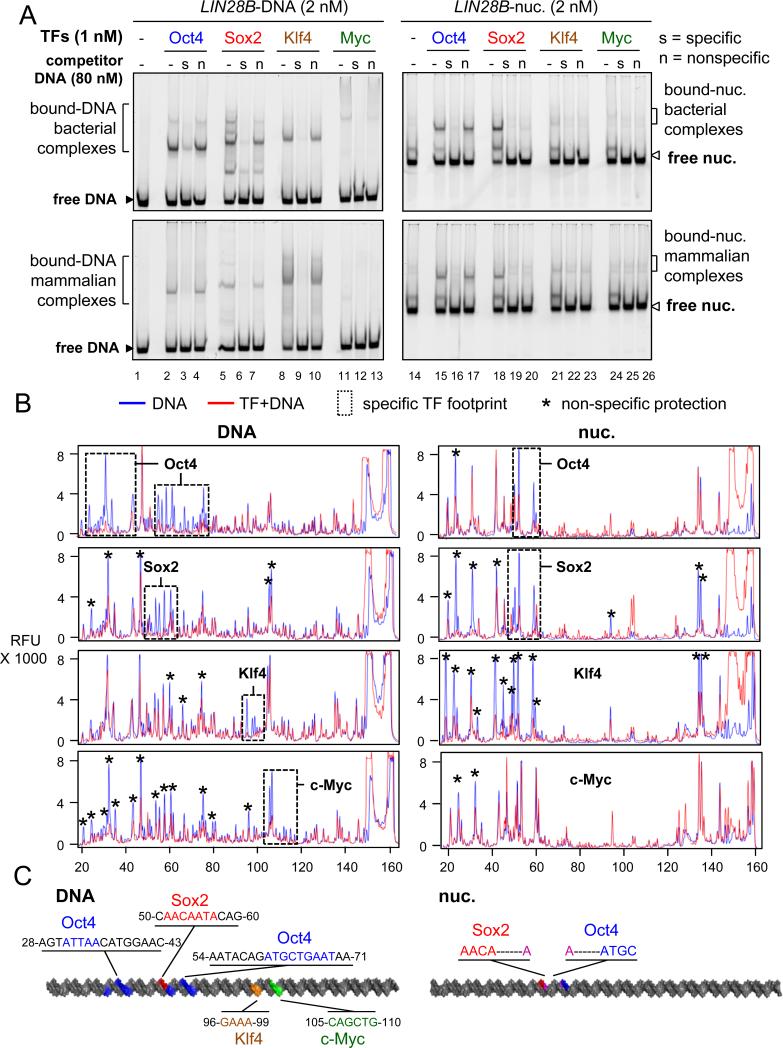
Figure 2. The contribution of non-specific binding to nucleosome targeting in vitro.
(A) Representative EMSA showing the affinity of recombinant O, S, K, M proteins (bact. top panels and mamm. bottom panels) to LIN28B-DNA (left panels) and LIN28B-nucleosome (right panels) in the presence of 40 fold molar excess of specific competitor ( “s” lanes) or non-specific competitor (“n” lanes) or absence of competitor (“-“ lanes). Competition assays showing the specificity of O, S, K, and M to their canonical DNA probes and to LIN28B DNA and nucleosome under lower titration of competitor is shown in Figure S2.
(B) DNase-I footprinting showing the protection of LIN28B-DNA (left panels) and LIN28B-nuc (right panels) in the absence (blue lines) or presence (red lines) of O, S, K, and M. Electropherograms of 5’-6FAM end-labeled LIN28B (top strand) oligonucleotides generated by DNase-I digestion of DNA (0.006 U) and nucleosomal DNA (0.06 U). Dashed boxes and stars represent specific and non-specific sites protected by O, S, K, and M, respectively.
(C) A cartoon representation of the 162 bp LIN28B DNA (left) and nucleosome (right) highlighting the binding sites of O, S, K, and M in vitro in blue, red, orange, and green, respectively, as measured by DNase-I footprining. The protected DNA sequences are indicated.