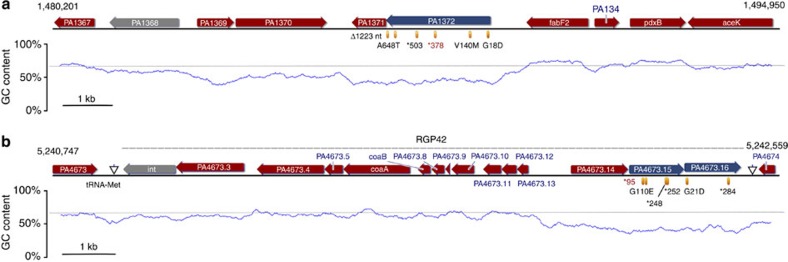
Figure 1. Genetic environment and mutations in PA1372 and PA4673.15-16.
Loss-of-function mutations in three recently acquired genes compensate for the cost of pNUK73 in PAO1. The schematic diagrams show the genetic environment of (a) gene PA1372, a putative helicase and (b) genes PA4673.15-16 two contiguous putative serine/threonine protein kinases. Mutations in these genes are responsible for compensatory adaptation to plasmid pNUK73 in P. aeruginosa PAO1. The reading frames for genes are shown as arrows, with the direction of transcription indicated by the arrowhead. PA1372 and PA4673.15-16 are shown in blue. Genes involved in genetic transposition or integration are shown in grey. PA1368 is a transposase belonging to the IS4 family and PA4673.2 (int) is the RGP42 phage integrase. The rest of the genes are shown in red. The chart below the sequence represents GC content (blue line), with the black dotted line indicating the median GC content of the PAO1 genome (67%). The dashed line above the sequence in panel B indicates the position of the prophage RGP42 with the arrows pointing at the insertion site next to a tRNA-Met coding sequence. Yellow ellipses indicate the positions of the mutations responsible for compensatory adaptation to pNUK73 and the changes in the predicted protein sequence are described below them (an asterisk indicates a premature stop codon and Δ indicates a deletion). We found each one of these mutations independently in different clones that have compensated for the cost of plasmid pNUK73 from different populations. The clones used in this study are those with the mutations highlighted in red. Whole-genome sequencing showed that these two mutant clones carry no other modifications compared with parental PAO1/pNUK73 apart from the compensatory mutation. The numbers in the left and right above the sequences indicate the genomic location in the P. aeruginosa PAO1 genome (NCBI taxonomy ID: 208964).