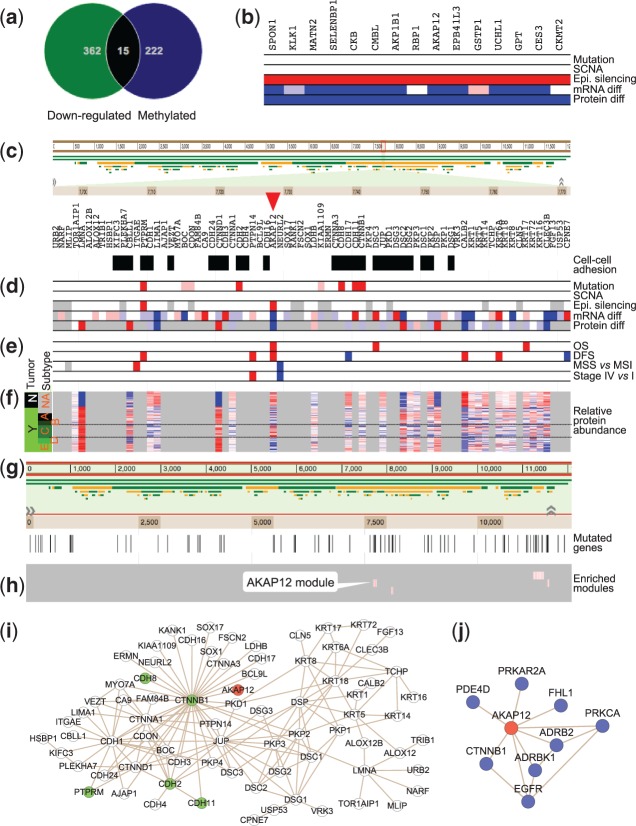
Fig. 4.
Prioritizing and interpreting epigenetically silenced genes. (a) Venn diagram analysis identified 15 epigenetically silenced genes that were also significantly down-regulated at the protein level in CRC. (b) Using the filtering feature, an omics snapshot for the 15 genes was created. (c) AKAP12 resides in a network module with 76 genes, and the module is significantly enriched with cell-cell adhesion genes. (d) The omics snapshot revealed five significantly mutated genes in this module. (e) The clinical relevance snapshot showed that higher mRNA level of AKAP12 was significantly associated with decreased OS and DFS time. (f) A detailed view of the proteomics data showed that compared with normal colon tissue, AKAP12 is down-expressed in most tumor samples except for subtype C tumors. (g) Non-random distribution of the 92 significantly mutated genes in the network. (h) These genes are significantly enriched in four network modules, including the AKAP12 containing module. The four modules are colored in pink and mutated genes in these modules are colored in red. (i) The involvement of AKAP12 in this module is primarily mediated through the interaction with CTNNB1. (j) All direct interaction partners of AKAP12