Fig. 1.
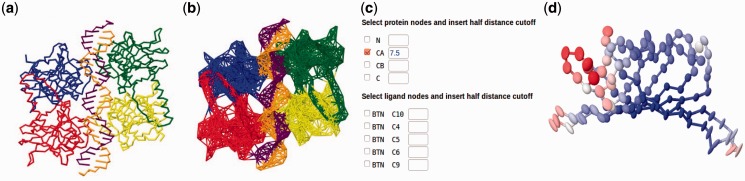
Some of the new features of ANM 2.0 web server: The new version is now applicable to nucleotide-containing structures. A snapshot of p53 bound to DNA (PDB 3Q05) is displayed by backbone stick representation (a) as well as network representation (b). Each nucleotide is represented by three nodes in this default setting, with a uniform 15Ȧ cutoff distance. (c) Versatility of the server for defining network nodes and assigning atom-specific interaction ranges. (d) Interactive representation of anisotropic ADPs as color-coded ellipsoids, supported by Jmol version 13.0.16. The ADPs computed for streptavidin (PDB 1STP) are shown. This PDB file contains no experimental data on ADPs