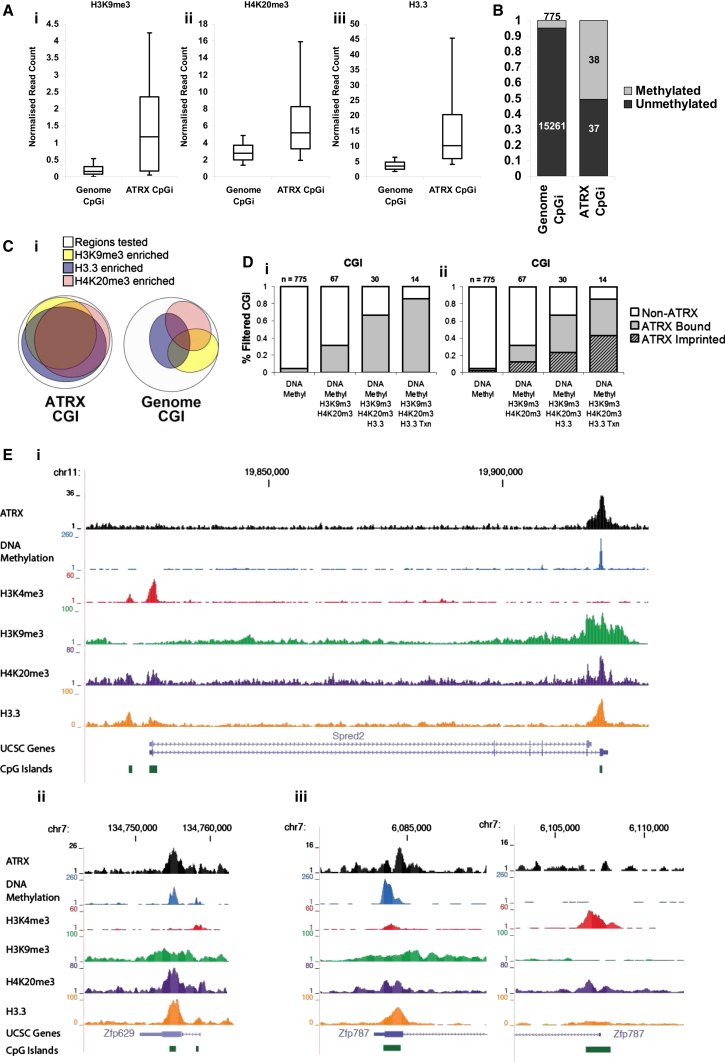
Figure 2.
ATRX-Bound CGI Are Enriched for Marks of Heterochromatin (H3K9me3/H4K20me3 and DNA Methylation) and H3.3
(A) Normalized read counts of (left) H3K9me3, (middle) H4K20me3, and (right) H3.3 ChIP-seq at ATRX-bound CGI relative to genomic CGI are shown. Boxes represent the 25th, median, and 75th percentiles; whiskers represent the 90th and 10th percentiles.
(B) Proportion of CGI that are methylated genome-wide in comparison to ATRX-bound CGI is shown.
(C) Overlapping enrichment profiles of H3K9me3, H4K20me3, and H3.3 under ATRX CGI compared to genomic CGI are shown.
(D) (Left) Proportion of CGI that are bound by ATRX following sequential filtering for DNA methylation, H3K9me3/H4K20me3 enrichment, H3.3 enrichment, and active transcription is shown. (Right) Proportion of CGI that are associated with imprinting and ATRX after sequential filtering is shown.
(E) Screenshots of the (top) Spred2 locus, (bottom left) Zfp629 locus, and (bottom right) Zfp787 locus show a typical promoter CGI marked by high H3K4me3 and mid-level H3.3 compared to an ATRX-bound intragenic CGI, which is enriched with marks of heterochromatin (DNA methylation, H3K9me3, and H4K20me3) and H3.3. See also Figure S2.