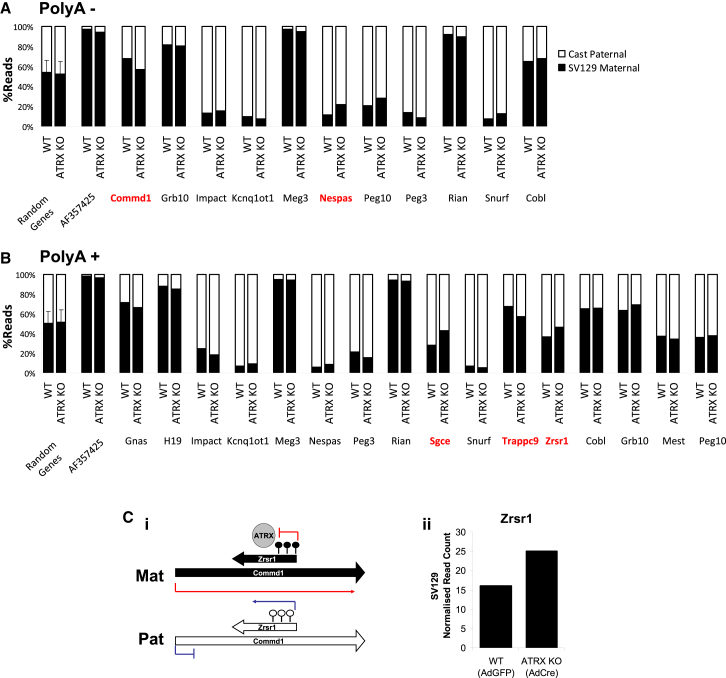
Figure 5.
Allelic Read Distribution at Imprinted Genes Is Disrupted in ATRX KO Cells
(A and B) RNA-seq of WT and ATRX KO ESCs show skewed distribution of reads at a number of imprinted genes in the (A) PolyA− and (B) PolyA+ fractions in WT cells. Genes where allelic distribution differed by >10% in ATRX KO compared to WT cells are indicated in red. Allelic distribution of reads across randomly chosen genes in the genome are shown for comparison. Error bars represent SD of allelic skewing across the random dataset.
(C) (Left) A schematic representation of the Zrsr1/Commd1 imprinted locus shows allelic expression and methylation patterns. (Right) Normalized read counts arise from the maternal SV129 Zrsr1 allele in WT and ATRX KO cells. See also Figure S6.