Figure 1.
mNET-Seq Methodology
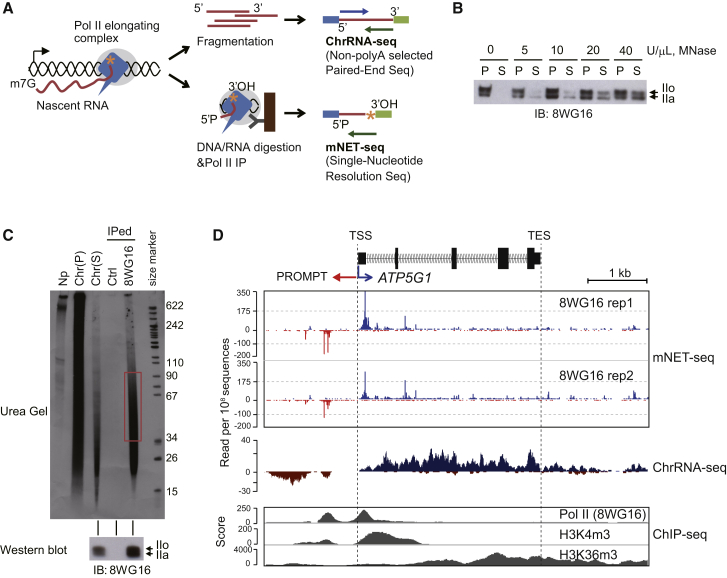
(A) ChrRNA-seq and mNET-seq strategies. Pol II (blue) elongating complex (gray circle) and associated nascent RNA (red line) in chromatin. Orange asterisk depicts the 3'OH of nascent RNA. For ChrRNA-seq (top), fragmented nascent RNA is subjected to directional paired-end deep sequencing. For mNET-seq (bottom), DNA and RNA are digested with MNase and Pol II-nascent RNA complex precipitated with Pol II antibody. Isolated RNA is deep sequenced, and the 3′ end nucleotide uniquely mapped on the human genome.
(B) Pol II release from insoluble chromatin DNA. Chromatin DNA was digested with increasing amounts of MNase. Western blot used 8WG16 Pol II antibody. P; pellet, S; supernatant. IIo and IIa indicate phosphorylated and unphosphorylated Pol II.
(C) Nascent RNA distribution in mNET-seq method. Nascent RNA was 32P-labeled by NRO reaction. Fractionated nascent RNA are nucleoplasm (Np), chromatin pellet (Chr (P)) and supernatant (Chr (S)). IP was with 8WG16 Pol II antibody. 35–100 nt RNA purified from gel (red box). IPed Pol II was detected by western blot (bottom).
(D) ATP5G1 mNET-seq. Two biological replicates of mNET-seq/unph using 8WG16 Pol II antibody. ChrRNA-seq shown as mNET-seq input. ChIP-seq (Pol II [8WG16], H3K4m3, and H3K36m3) data are from ENCODE project data sets (Consortium et al., 2012).