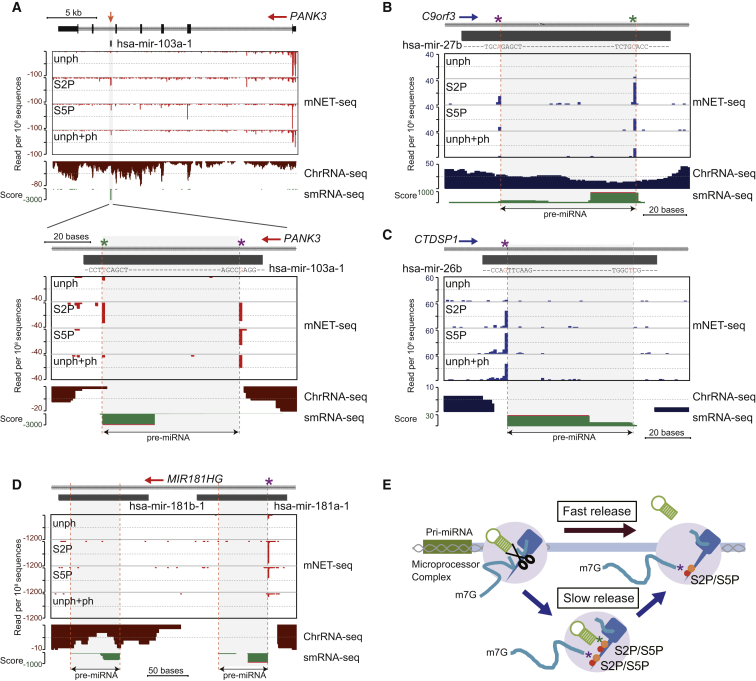
Figure 5.
Pre-miRNA Biogenesis from Protein-Coding Gene Introns
(A–D) mNET-seq with different Pol II antibodies versus ChrRNA-seq over intronic pre-miRNAs. (A) mNET-seq data on PANK3 with magnified view over hsa-mir-103a-1 denoted by a black rectangle. The pre-miRNA is indicated by an orange arrow (top). Three other pre-miRNA are also shown: hsa-mir-27b (B), hsa-mir-26b (C), and hsa-mir181a/b-1 (D). Drosha cleavage sites are identified by dashed orange lines, and asterisks indicate frequent cleavage sites (5′ end, purple; 3′end, green). Small RNA-seq data are shown below (green).
(E) Model of co-transcriptional pre-miRNA biogenesis. Pre-miRNA DNA and hairpin RNA are shown in green. Co-transcriptional Drosha cleavage (scissors) and spliceosome (gray) shown with 3′ ends of cleaved RNA (purple asterisk) and pre-miRNA (green asterisk) tethered to phosphorylated CTD. Pre-miRNA release may occur from the transcription complex, fast (dark red arrow) or slow (blue arrows).