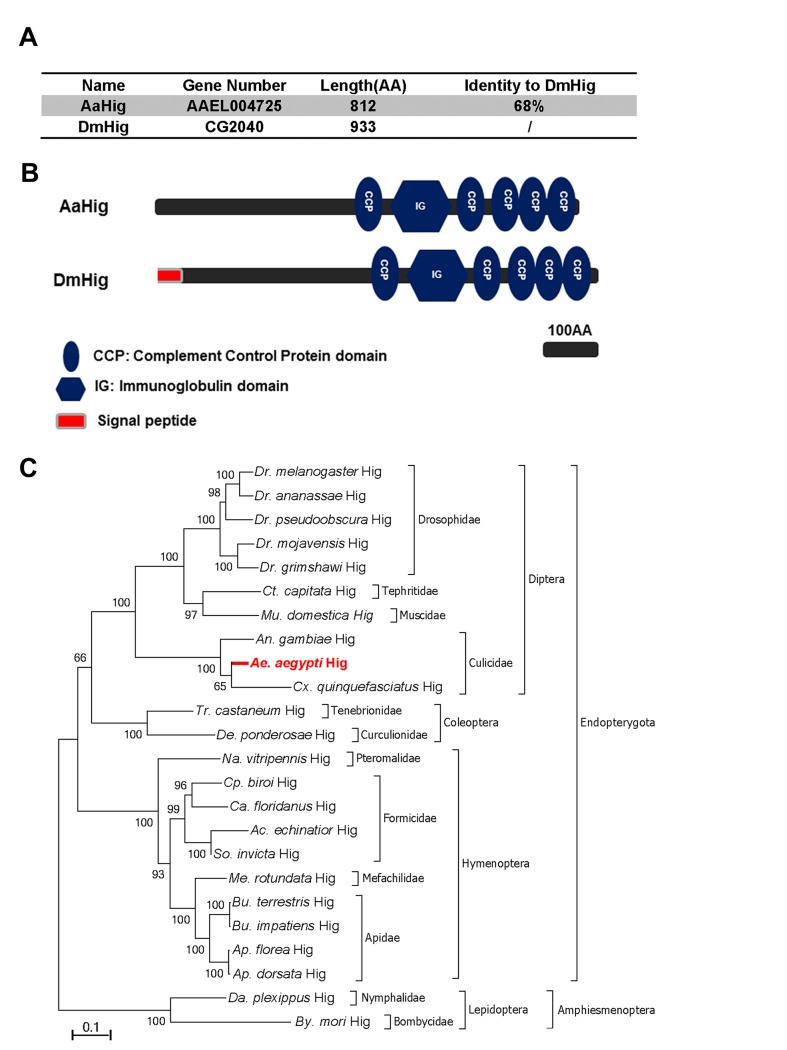
Fig 1. Bioinformatic comparison and phylogenetic analysis of Hikaru genki (Hig) in insects.
(A) Percentage of amino acid identity between AaHig and Drosophila melanogaster Hig (DmHig). (B) Schematic representation of AaHig and DmHig. The functional modules were predicted using the SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi?GENOMIC=1) and Pfam (http://pfam.sanger.ac.uk/) websites. (C) Unrooted phylogenetic tree of insect Higs. The tree was constructed using the neighbour-joining (NJ) method based on the alignment of insect Hig protein sequences. The bootstrap values of 5000 replicates are indicated on the branch nodes. Drosophila spp. (Dr.); Ceratitis spp. (Ct.); Musca spp. (Mu); Anopheles spp. (An.); Aedes spp. (Ae.); Culex spp. (Cx); Tribolium spp. (Tr.); Dendroctonus spp. (De.); Nasonia spp. (Na.); Cerapachys spp. (Cp.); Camponotus spp. (Ca.); Acromyrmex spp. (Ac.); Solenopsis spp. (So.); Megachile spp. (Me.); Bombus spp. (Bu.); Apis spp. (Ap.); Danaus spp. (Da.); Bombyx spp. (By.).