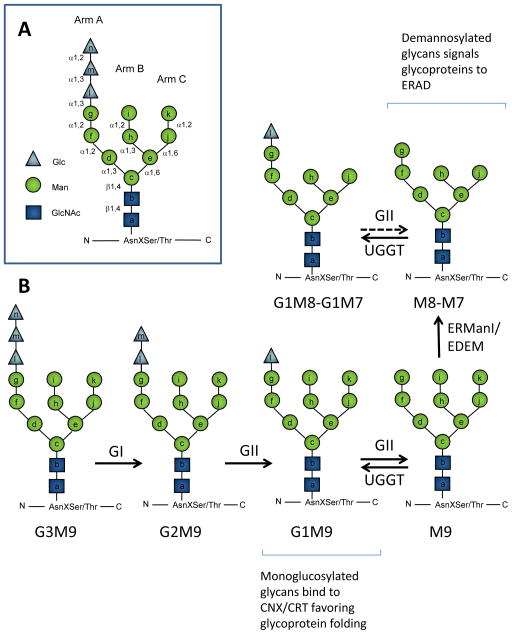
Figure 1. Processing of N-glycans in the ER.
A. Oligosaccharide Glc3Man9GlcNAc2 (G3M9) is transferred to Asn residues on nascent polypeptides by oligosaccharyltransferase. Lettering a–n indicates the order of addition of the monosaccharides during in vivo synthesis of the Dolichol-PP-Glc3Man9GlcNAc2 precursor. Arm A, B and C indicate the oligosaccharide branch. During biosynthesis residues a–g are added on the cytosolic face of the ER membrane from nucleotide-sugar precursors, while residues h–n are added from Dol-P-Glc or Dol-P-Man precursors after the oligosaccharide has flipped across the membrane. B. After glycan transfer to proteins, Glucosidase I (GI) removes glucose n, Glucosidase II (GII) removes glucose m and l, and UDP-Glc:glycoprotein glucosyltransferase (UGGT) adds glucose l. ER mannosidases may remove mannoseI and k. Monoglucosylated N-glycans are able to interact with ER lectins calnexin (CNX) and/or calreticulin (CRT). ER mannosidase I (ERManI) (MNSI in yeasts) acts as a timer of permanence of glycoproteins in the ER and removes residue I generating M8B. Subsequently, ERManI or other ER mannosidases (probably EDEM in mammals or Htm1in yeasts) may remove residue k, generating M7. Demannosylated glycans constitute the ERAD signal. Dotted lines are used to indicate that GII activity is reduced toward demmanosylated species.