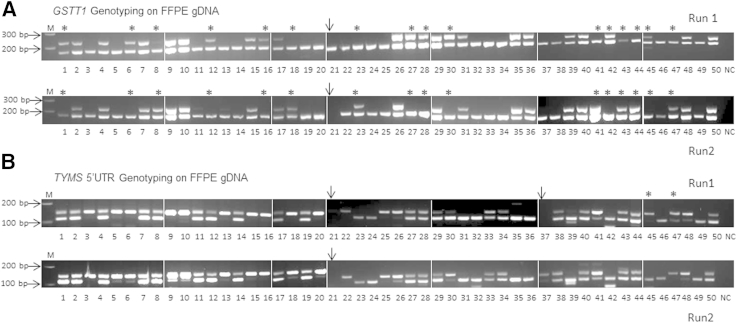
Figure 1.
GSTT1 and TYMS 5′ untranslated region (UTR) genotyping on formalin-fixed, paraffin-embedded (FFPE) samples. A:GSTT1 genotyping on 50 FFPE samples. Samples 1 to 30 are from the MAGIC cohort and 31 to 50 from the JUST cohort. Complete absence of the GSTT1 product (top panel) in the presence of an ACTB amplicon (bottom panel) was interpreted as a homozygous deletion (GSTT1 *0/0 null genotype). The presence of a GSTT1 product was interpreted as GSTT1 positive. Samples labeled with stars exhibit discrepant results between two independent runs. Sample 21 (arrow) failed in the second run and was excluded from the analysis. The concordance rate between the two runs was 69.4% for FFPE DNA. B:TYMS 5′-UTR genotyping on 50 FFPE samples. Sequence repeats in the TYMS 5′-UTR enhancer region can be classified into 2R/2R, 2R/3R, and 3R/3R genotypes. Samples labeled with stars exhibited discrepant results between the two independent runs. Sample 21 (arrow) failed in both runs, and sample 37 failed in the first run. The concordance rate between the two runs was 95.8% for FFPE DNA. gDNA, genomic DNA; M, 100-bp DNA ladder; NC, negative control.