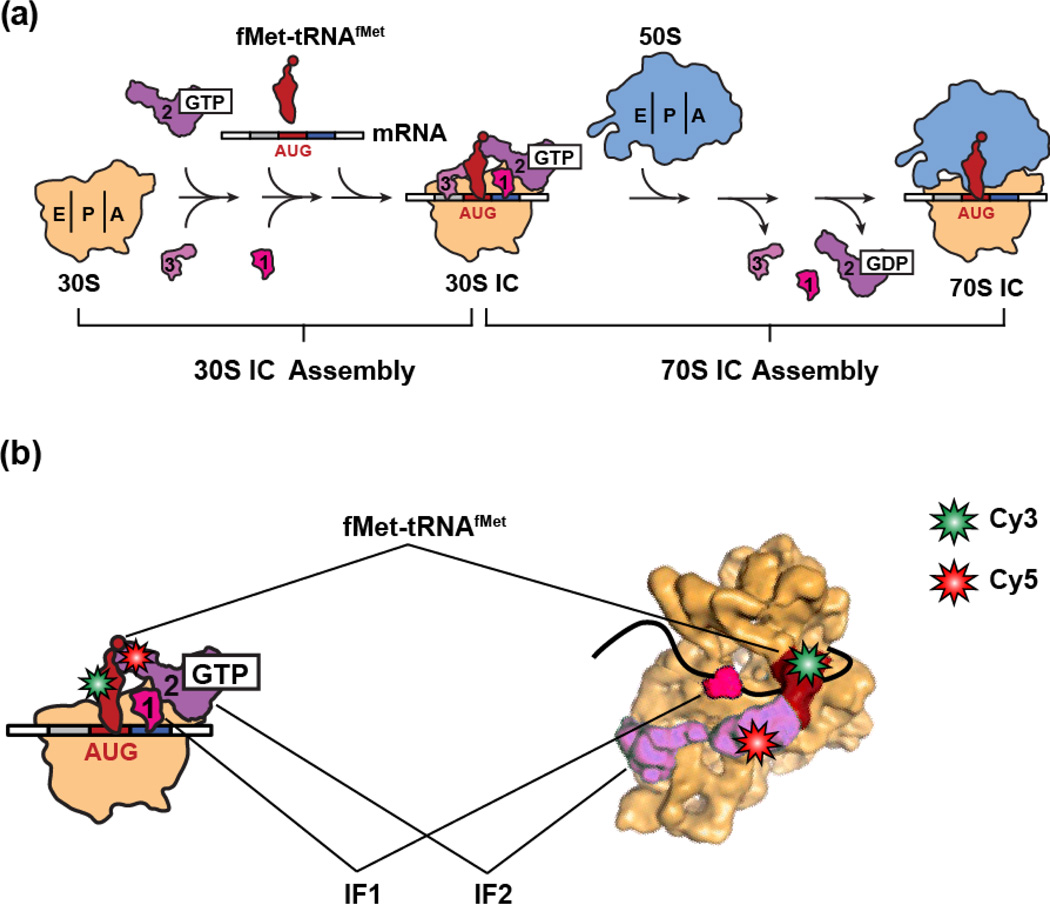
Figure 1.
(a) Cartoon representation of the canonical translation initiation pathway. During the first major step of translation initiation, the IFs bind to the 30S subunit and collaboratively increase the rate and accuracy of fMet-tRNAfMet selection and decoding of the start codon within the P site of the 30S subunit, resulting in the assembly of a 30S IC at the start codon of the mRNA to be translated. During the second major step, the 50S subunit joins to the 30S IC, the IFs are released from the resulting 70S IC, and fMet-tRNAfMet is positioned into the peptidyl transferase center of the 50S subunit, resulting in the assembly of an elongation-competent 70S IC. (b) A cartoon representation (left panel) and cryogenic electron microscopy (cryo-EM) structural model (right panel) depicting the IF2-tRNA smFRET signal used to monitor the conformational dynamics of the IF2•tRNA sub-complex within 30S ICs. IF2 was labeled with a Cy5 FRET acceptor at a cysteine residue that was engineered into domain IV of IF2 and fMet-tRNAfMet or variants thereof were labeled with a Cy3 FRET donor within their central fold, or “elbow”, domains. The figure depicting the cryo-EM structural model was adapted from a figure published in Simonetti et al., 200817.