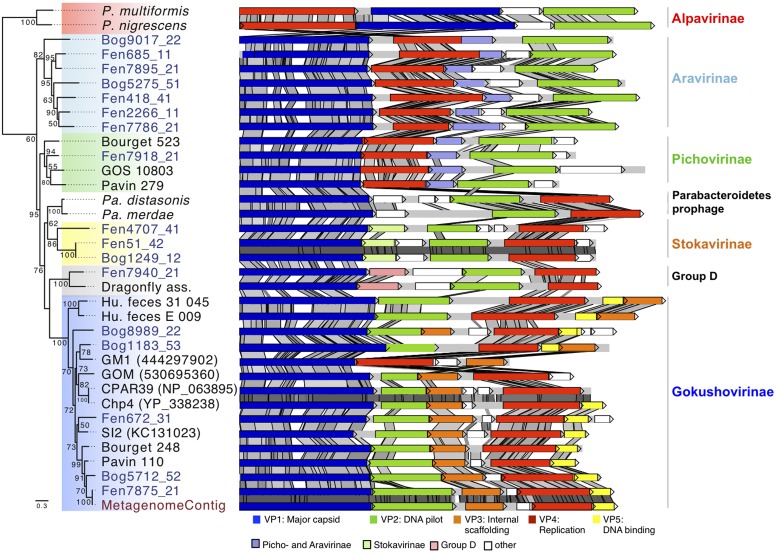
FIGURE 3.
Major capsid protein phylogeny and genome structure of major subfamilies of Microviridae bacteriophages. The affiliation of the peat genomes to Microviridae is shown by major capsid protein phylogeny (Maximum likelihood, 252aa positions, 1000 iterations, JTT+G model). Bootstrap values above 50% are indicated at the nodes. The genome structures of the assembled viral peat genomes (blue) were compared to other Microviridae genomes. Pairwise comparison (TBLASTX) was visualized with ACT (Carver et al., 2005). Gray shading indicates the level of similarities. Homologous genes specific for some subfamilies are color-coded as indicated in the legend. P. multiformis, Prevotella multiformis (WP_007368517); P. nigrescens, Prevotella nigrescens (WP_023924568); Pa. distasonis, Parabacteroides distasonis (ZP_17317332); Pa. merdae, Parabacteroides merdae (WP_022322420). Dragonfly, Dragonfly associated phage (YP_006908226); GOS_10803 (ECU79166); GM1 (444297902); GOM (530695360); CPAR39 (NP_063895); Chp4 (YP_338238); SI2 (KC131023); Metagenome: genome assembled from the metagenome.