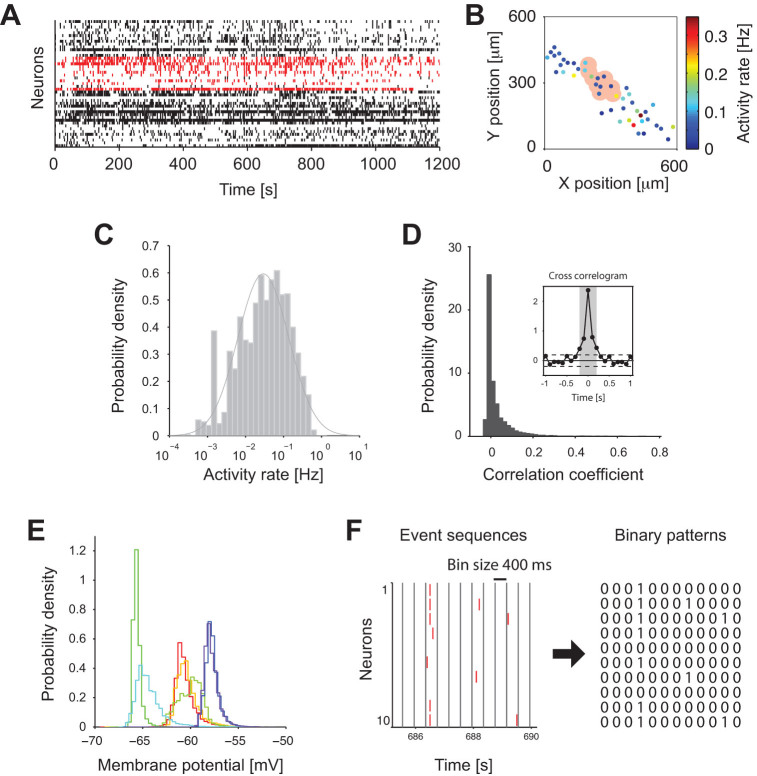
Figure 1. Ensemble activity of CA3 putative neurons detected by Calcium imaging.
(A) Ensemble activity of 45 neurons from a single hippocampal slice. Small vertical ticks indicate events detected from calcium imaging signals. Ensemble activity of an example group of 10 neurons is marked in red. (B) Spatial distribution of neurons in the CA3 area of the slice in A. Each filled circle represents a position of a neuron. The color indicates activity rate of each neuron. The pink area corresponds to the example group highlighted in A. (C) Distribution of activity rates from neurons in all 20 slices. Solid line is a fitted log-normal distribution. (D) Distribution of correlation coefficients calculated from the event sequences (within a 100 ms window) from all the pairs of neurons in 1000 neighboring groups from 20 slices. The inset shows an average cross-correlogram from all the pairs of neurons. Dashed lines indicate ± 2 SD of the correlogram at 1–2 sec lags. The gray shading (−0.2 ms to + 0.2 ms) indicates the interval where the correlgoram exceeded the dashed lines. (E) Distributions of membrane potentials recorded from neurons (n = 7) in hippocampal slice cultures under the same condition as described in Methods. Different colors indicate different neurons. In all cases, the densities of the membrane potentials were characterized by a unimodal profile. (F) Construction of binary patterns from event sequences. The event sequences are binned using a window of 400 ms. In each bin, we denote ‘0’ if there is no event, and ‘1’ if there is at least one event.