Figure 1.
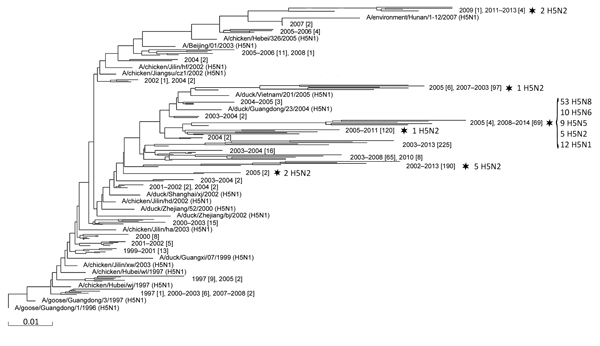
Phylogenetic tree showing the evolutionary history of the hemagglutinin (HA) proteins of novel highly pathogenic avian influenza (HPAI) H5 HA subtype viruses. By using MUSCLE (9), we aligned the coding region sequences for 89 HPAI H5 HA subtype viruses, excluding H5N1, with those for 850 H5N1 HA viruses representing all HPAI H5N1 clades (1); the 89 H5 HA sequences were identified in the NCBI Influenza Virus Resource (10) and the GISAID EpiFlu Database (http://www.gisaid.org). A phylogenetic tree was constructed by using the PHYLIP Neighbor Joining algorithm using the F84 distance matrix (http://www.ncbi.nlm.nih.gov/genomes/FLU/DatasetExplorer/fluPage.cgi?pageInclude=References.inc#PHYLIP). The number of sequences present in a branch is indicated between brackets. Stars indicate the branches that contain subtypes other than H5N1. The genotypes (H5N2, H5N5, H5N6, and H5N8) and their numbers of occurrence in a particular branch are indicated at right. Scale bar indicates evolutionary distance (nucleotide substitutions per site). Details for GISAID-derived sequences are shown in the Table.
